Main Content
Project 1: A Correlative Metabologenomics Approach for the Discovery of Specialized Metabolites in Marine Fungi

Secondary or specialized metabolites (SMs) remain the most important source of drug leads. Research in the last decades highlights the great genetic potential and increasing importance of fungi for finding bioactive and novel SMs. However, this potential is far from being exhausted, especially, SMs from understudied sources like marine-derived fungi. The main reasons are limited sequencing data for the latter and the presence of cryptic/silent genes, potentially making up 90% of all biosynthetic gene clusters (BGCs). To fill this gap and to unravel SMs as well as their genetic potential, we plan to sequence the genomes and analyze transcriptomes of less studied strains, 38 Phoma and 12 Acremonium, from diverse ecological marine niches and locations, which are mostly associated with other organisms like sponges and macroalgae. We expect to find novel bioactive SMs, intriguing enzymes and catalyzed reactions from these strains.
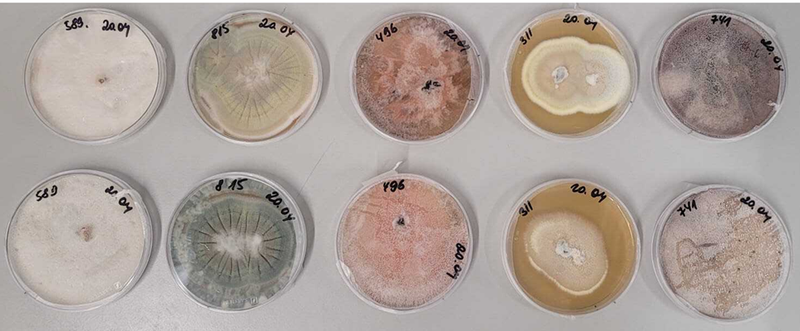
Selected marine Phoma sp. from diverse ecological niches.
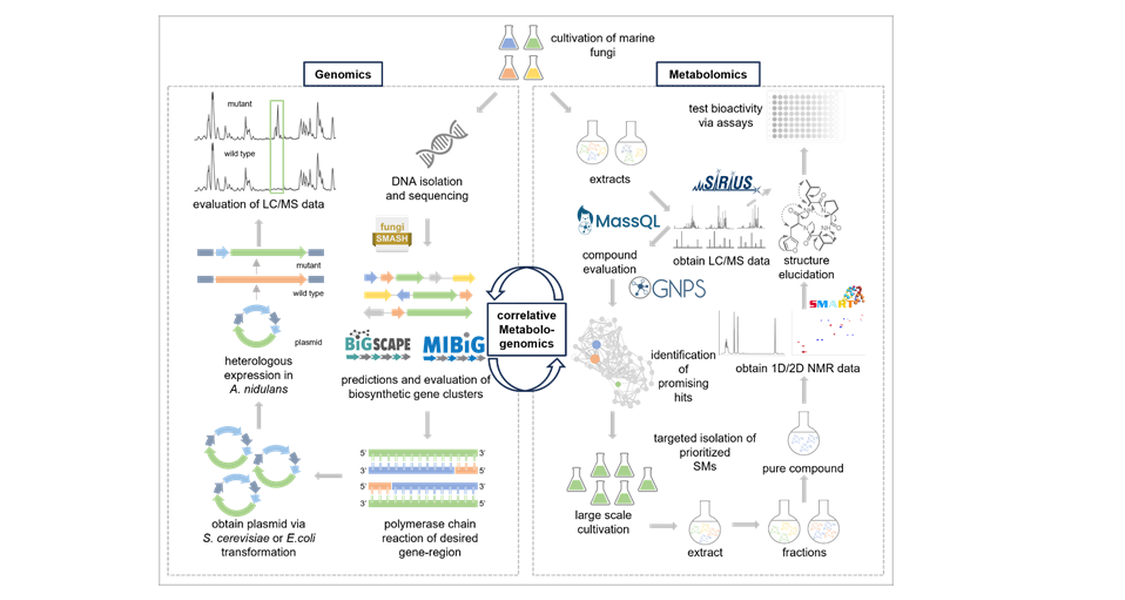
Meanwhile, the genome sequences are also important for other research fields, e.g. studying on their relationships to hosts and environments. Correlative metabologenomics allows the comparison of fungal metabolites produced with the predicted biosynthetic gene clusters from the associated genome. Therefore, known natural products will be detected from LC/MS data and clustered and dereplicated using the GNPS framework with its integrated database. The compounds will then be further investigated using in-silico frameworks such as SIRIUS. Fungal BGCs will be identified by fungiSMASH and clustered by BiG-SCAPE using the MIBiG database for annotation of known BGCs.
Recently, we developed a MassQL-Integrated Molecular Networking Approach for the Discovery and Substructure Annotation of Bioactive Cyclic Peptides from the marine sponge-derived fungus Stachylidium bicolor 293 K04.
Project 2: The Role of Marine Fungi in Tackling Plastic Waste: A Multiomics Approach
Marine-derived fungi are widely recognised as prolific producers of bioactive natural products.For survival in ecological niches (high salt content, tropical or arctic temperatures, respectively, endosymbiotic relationships), a multitude of enzymatic reactions, which can be both highly specific and complex in nature, occur within these organisms. In consideration of the enormous annual global plastic production, which inevitably results in a problematic accumulation of plastic in the oceans, there is potential for investigation of the ability of fungal enzymes to degrade plastic polymers. In this study, we want to test whether native organisms of the marine environment, such as marine-derived fungi, can offer a fruitful source for plastic degradation.
It has previously been demonstrated that a number of marine fungi collected in the waters off the Hawaiian island of O'ahu have exhibited a visible clearance effect on polyurethane (PU)-containing agar plates, hinting towards a degradation process.To procure more detailed information
regarding this process, the present study is implementing a multiomics approach, including genomics, metabolomics, and transcriptomics experiments .
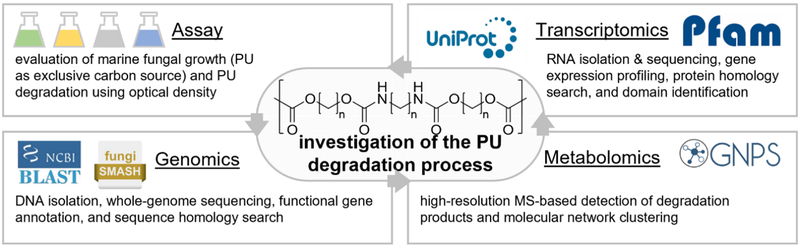
This approach is designed to facilitate the identification of the implicated enzymes and enable their correlation with the detected metabolic products. Therefore, the genomes of the investigated marine fungi were analyzed to ascertain the possible presence of any enzymes that have previously been
identified as capable of degrading plastic polymers, as documented in the PAZy Database. In addition, high-resolution mass spectrometry facilitates the identification of degradation products through the utilization of MS-based molecular networking, which employs a clustering approach to organize features according to their analogous fragment patterns.[5] Finally, the exploration of overexpressed genes during RNA sequencing analysis ultimately led to the correlation of specific modifications in the molecular structure of the PU polymer to specific present enzymes and their associated functions.
In summary, the results of the study provide valuable insights into the process of enzymatic degradation of PU by marine-derived fungi. Moreover, the findings of this research establish a solid foundation for the development of novel strategies for the management of plastic waste.
Projects 1 + 2 are investigated by Tim Berger.

Project 3: MS-integrated Reactivity-Based Genome Mining:
A Novel Approach for Natural Product Discovery
Marine fungal specialized metabolites (SMs) are an important source of bioactive small molecules. However, their discovery is hampered by frequent re-isolation of known compounds, low or silent expression of biosynthetic gene clusters (BGCs), and the difficulty of linking BGCs to their corresponding metabolites.
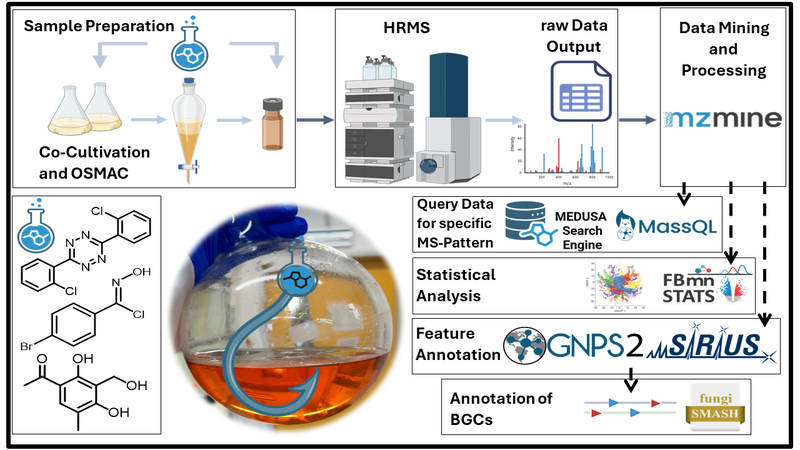
Here, we introduce MS-integrated Reactivity-based Genome Mining (MS-RGM). This approach employs reactive targeted and untargeted chemical probes to capture (i) SMs with defined functional groups and (ii) volatile metabolites and biosynthetic intermediates using broadly reactive probes. The resulting adducts are detected through MassQL-based MS queries and subsequently prioritized for isolation and structural elucidation (Fig. 1). In addition, silent BGCs can be activated by ecological interactions in co-culture, OSMAC strategies, or probe-induced chemical stress. Compared to classical genome mining, MSRGM enables more efficient BGC–SM connections and expands accessible chemical space by generating “unnatural” pseudo-SMs for bioactivity screening.
This project is investigated by Bastian Brand
