Main Content
Highlights 2025
- I am very honored that I received the Young Investigator (Horst-Böhme) Award of the German Pharmaceutical Society (DPhG). Press release Uni Marburg
- Let's go to VAAM 2025 in Berlin. We are thrilled to present our research with two talks and one poster presentation.
- Thursday Sep 24 Amira Naimi SL8: 12:15-12:30 pm Native metabolomics-guided Discovery of a Cell-Permeable Gαi-Inhibitor"
- Friday Sep 25 SL18: 11:45 – 12:00 Tim Berger, "The Role of Marine Fungi in Tackling Plastic Waste: A Multiomics Approach"
- Poster P47: Bastian Brand "MS-integrated Reactivity-Based Genome Mining: A Novel Approach for Natural Product Discovery"
- I feel very honored to have received the Beal Award 2025 from JNP-Editor Brad Moore @ ASP 2025 in Grand Rapids, Michigan. Please find the interview here.
- Finally, the MassQL main manuscript appeared in Nature Methods, a "MassIVE" community-effort with more than 50 co-authors led by Prof. Ming Wang (UCR). Congrats, Ming! Tim, already applied this pattern-based query language last year for the annotation and classification of cyclic peptides at the substructure level. These innovations are tightly integrated into our analytical workflows and represent a new level of hypothesis-free discovery (J. Nat. Prod.). Tim's Paper was instrumental in validating the platform’s utility for real-world discovery workflows and was recently honored with the Jack L. Beal Award 2025 for the best JNP-paper in the year 2024. Fantastic job, Tim! Additionally, it was highlighted in a research briefing and significantly contributed to the successful revision of the Nature Methods article above, where we also provided an application use-case (led by Prof. Timo Niedermeier, FU Berlin) identifying putative analogs of the “eagle killer toxin AETX“ in > 230 millions of public MS/MS spectra.
Highlights 2024
- In his first year Bastian already contributed to a publication where he applied AI-NMR tools in a highly interdisciplinary team with physicists, philologists, and experts for pharmaceutical history to subclassify the plant resin “elimi” that has recently be described to be involved in ancient Egyptian mummy embalming (Arch. Pharm). Way to go Basti!
- Everyone did a fantastic job during the poster presentations. We are very happy and proud to share that Amira's poster won one of few GA-awards among almost 1000 scientific posters. Awesome job Amira!

- We are looking forward to present our research at ICNPR 2024 in Kraków. If you want to learn more about our exciting research ranging from marine fungi over G-proteins to Ancient Egyptian artifacts, please stop at posters S1.P139 (Tim Berger), S1.P274 (Bastian Brand), and S3.P100 (Amira Naimi).
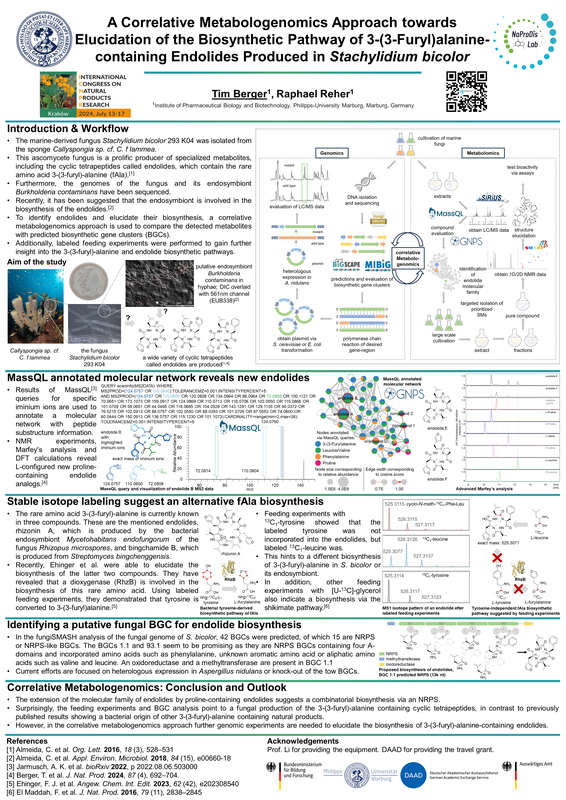


- Tim Berger developed a method that allows for substructure annotations of molecular networks by applying MassQL, boosting MS2-based annotation rates for peptides, guiding the isolation of new specialized metabolite from marine fungi (or their endosymbionts?) Have a look at our first research paper :-) Congrats Tim!

We are very happy that (almost licensed) pharmacist Bastian Brand joined our group. Welcome to the Natural Products Discovery Lab, Bastian!
Highlights 2023
- The NaProDis Lab and the FunctionalMetablomics Lab joined forces for a review on mass spectrometrical approaches to study protein-metabolite interactions. Great job Amira!
- Ein Bericht über den diesjährigen Betriebsausflug bei der Engelhard Arzneimittel GmbH & Co. KG, verfasst von Doktorandin Amira Naimi und Prof. Reher, ist in der Deutschen Apotheker Zeitung erschienen. Vielen Dank noch einmal für die spannende Führung und Organisation von Sonja Hiemenz!

- A great comunity effort covering synergies between omics-based natural product discovery and computational drug design "Artificial intelligence for natural product drug discovery http://bit.ly/488vLOB" was published in Nature Reviews Drug Discovery (IF 112) and received astonishing resonance in the media, (currently rankend 2nd among 55 tracked articles of a similar age in Nature Reviews Drug Discovery, link to altmetric scores).
Highlights 2022
- It was a great "Day of Pharmacy" at Marburg University where my colleagues an I were honored to give our Inaugural Lectures

- We are very happy that Sina joined or group in October 2022. Welcome to the NaProDis-Team :-)

- We are delighted to announce that our research about native metabolomics published in Nature Communications was highlighted in press releases of the Philipps-Universität Marburg and the University of Tübingen (english version)