Main Content
2) Deciphering the function of LDs in HCV replication and in other infectious diseases
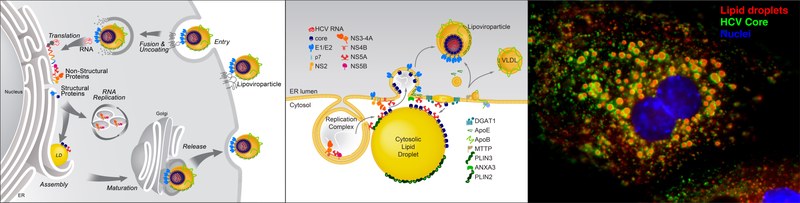
Model of HCV replication and morphogenesis at lipid droplets (Modified from Herker and Ott, J. Biol. Chem., 2012). HCV core localizes to lipid droplets in primary hepatocytes.
HCVs enveloped infectious lipoviroparticles contain a single (+) stranded RNA genome and are tightly associated with lipoproteins and neutral lipids. The virus replicates in the cytoplasm of human hepatocytes and encodes for one polyprotein that is cleaved into 10 proteins, the three structural proteins (the nucleocapsid core and two envelope glycoproteins E1 and E2), the viroporin p7, and six non-structural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The viral RNA is replicated by RNA replication complexes within ER-derived membrane structures termed the membranous web, which is detergent-resistant due to the presence of lipids that are associated with lipid microdomains, namely cholesterol and sphingolipids. Importantly, inhibition of cholesterol and sphingomyelin biosynthesis suppresses viral RNA replication in cells. We recently determined the lipid composition of HCV-infected cells and subcellular organelles and could link fatty acid elongation and desaturation to efficient viral replication (Hofmann et al., Biochim. Biophys. Acta, 2018). Most likely, specific membrane fluidity and curvature is required for the formation of the RNA replication vesicles. This membranous structure is thought to shield replication centers from detection by pattern recognition receptors and contains single-, double-, and multi-membrane vesicles as well as cytosolic LDs that are the putative site of viral assembly (Herker and Ott, Trends Endocrinol. Metab., 2011, Herker and Ott, J. Biol. Chem., 2012).
Our finding that functional LD biogenesis is required for efficient viral progeny production highlighted the importance of these host organelles for HCV replication (Herker et al., Nat. Med., 2010). Mechanistically, translocation of both core and NS5A to LDs requires triglyceride biosynthesis as inhibitors of diacylglycerol acyltransferase-1 (DGAT1) impair trafficking to LDs and subsequent HCV assembly (Herker et al., Nat. Med., 2010, Camus et al., J. Biol. Chem., 2013). HCV core protein also reduces LD turnover at the surface of LDs, thereby inducing steatosis in cultured cells and in murine livers (Harris et al., J. Biol. Chem., 2011, Camus et al., J. Biol. Chem., 2014). In our quantitative LD proteome analysis we observed that HCV disconnects LDs from their normal metabolic function and regulation and identified ANXA3 as a novel host factor for HCV maturation (Rosch et al., Cell Rep, 2016). Perilipin-2 (PLIN2/ADRP) is the major coat protein of LDs in hepatocytes. We recently investigated the consequences of PLIN2 deficiency on LDs as well as on HCV infection and found that PLIN2-deficient cells harbor lipid droplets trapped in double membrane sacs that do not support infectious HCV particle production (Lassen et al., J. Cell Sci., 2019).