Main Content
Elucidate impaired desmosomal Function in inflammatory Model diseases of Skin
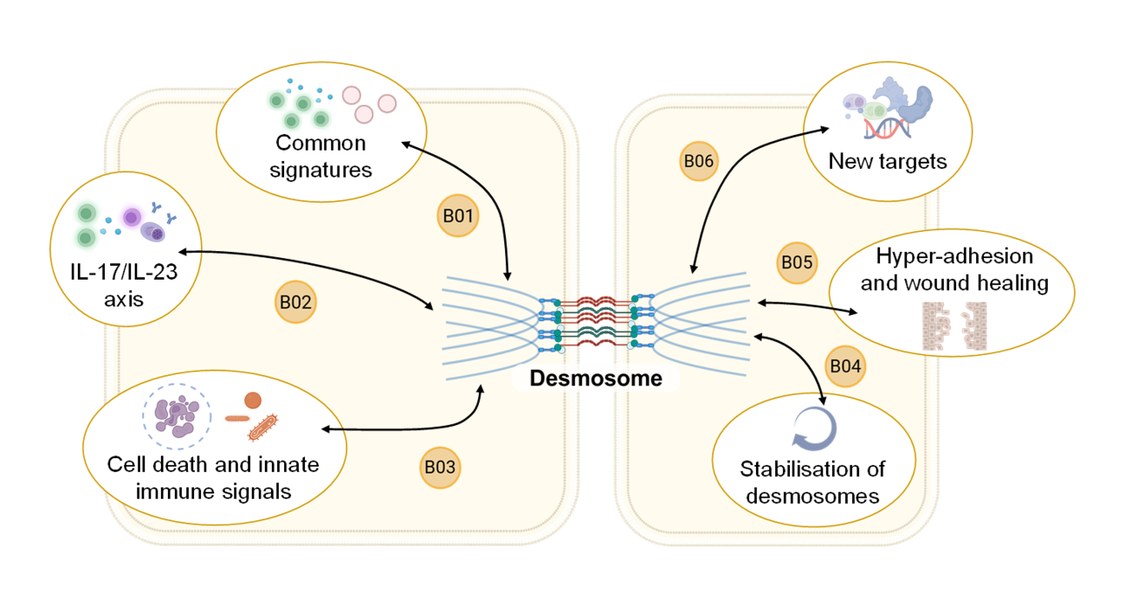
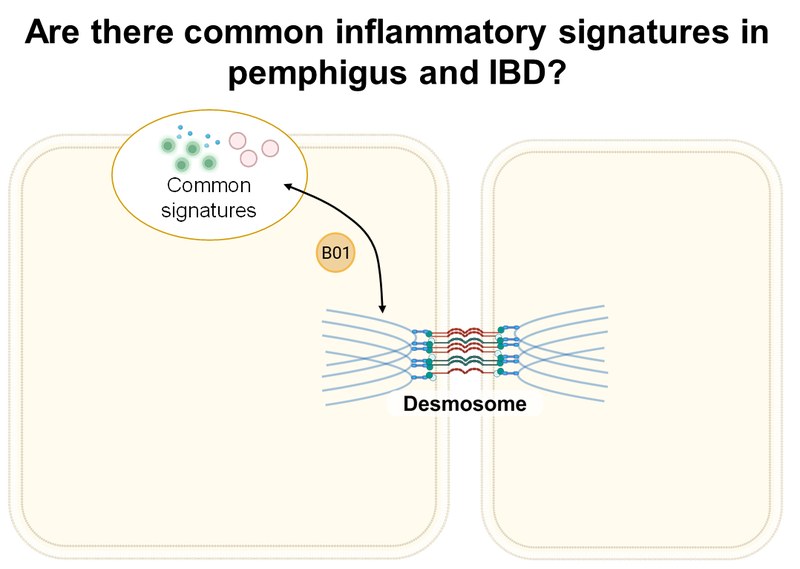
Inhalt ausklappen Inhalt einklappen B01 - Inflammatory signature in pemphigus
Michael Hertl, Department of Dermatology and Allergology, University of Marburg
Anna Lena Jung, iLung-Institute for Lung Research, University of Marburg
Project: B01 integrates extracellular vesicle (EV) profiling with functional T cell analyses to investigate their influence on epithelial skin structure and function in pemphigus vulgaris (PV). Using patient samples, in vitro and ex vivo organoid models, single-cell technologies, and a humanized PV model, the project aims to uncover core mechanisms of T cell processes in PV. Using a comparative approach with inflammatory bowel diseases (IBD), we aim to define common and tissue-specific pathways of immune-epithelial cell communication.Inhalt ausklappen Inhalt einklappen B02 - Regulation and modulation of T/B cell interactions, and implications of the IL-17/IL-23 axis in pemphigus
Christian Möbs, Department of Dermatology and Allergology, UMR
Magdalena Huber, Institute for Systems Immunology, UMR
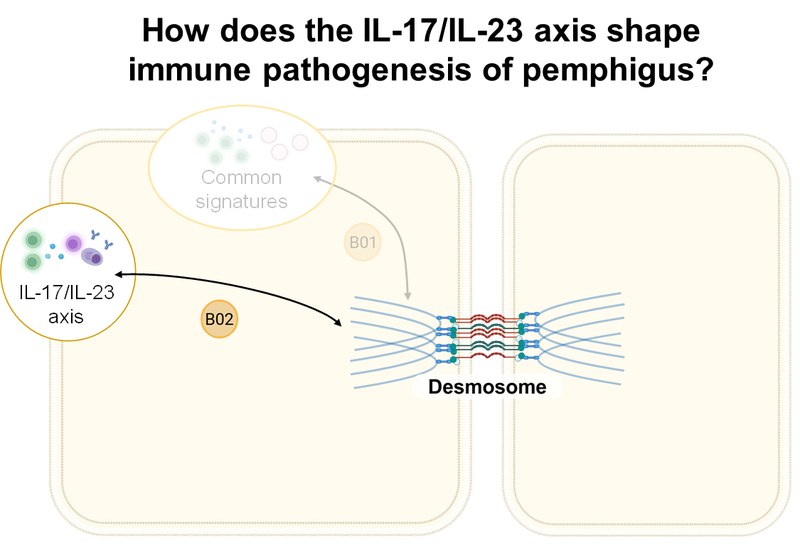
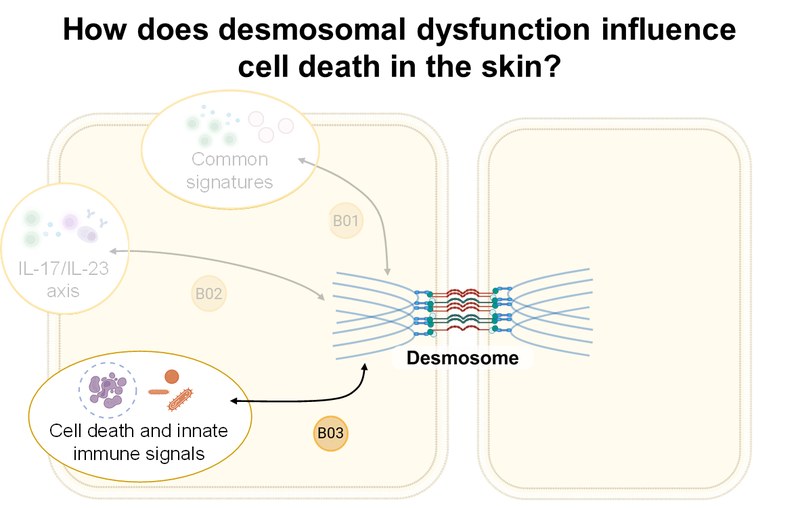
Project: Using high-dimensional single-cell technologies, accompanied by OLINK serum proteomics and tissue histology, B02 investigates (i) direct effects of type 17 cells and their secretome, (ii) indirect effects via the activation of B cells to autoantibody production by CXCR5+ follicular Th cells (Tfh) and CXCR5– peripheral Th cells (Tph), and (iii) altered counter regulation by Treg cells in the pathogenesis of pemphigus. These findings are validated in preclinical mouse models of PV.Inhalt ausklappen Inhalt einklappen B03 - Cell death and innate immune response caused by desmoglein dysfunction
Amir Yazdi, Department of Dermatology and Allergology, RWTH Aachen University
Felix Lauffer, Department of Dermatology and Allergology, LMU Munich
Project: B03 aims to elucidate the mechanisms of desmosomal adhesion loss in pemphigus vulgaris (PV), bullous impetigo (BI), and Darier's disease (DD). Using RNAseq and immunofluorescence of tissue samples, disease-specific and overlapping gene signatures in the three entities will be analyzed. Using CRISPR/Cas9, relevant signaling pathways (necroptosis, apoptosis, pyroptosis, ferroptosis) will be blocked. 3D skin equivalents will be used, among other methods.
Open Position available! (check for link)
Inhalt ausklappen Inhalt einklappen B04 - Stabilisation of keratinocyte adhesion in pemphigus
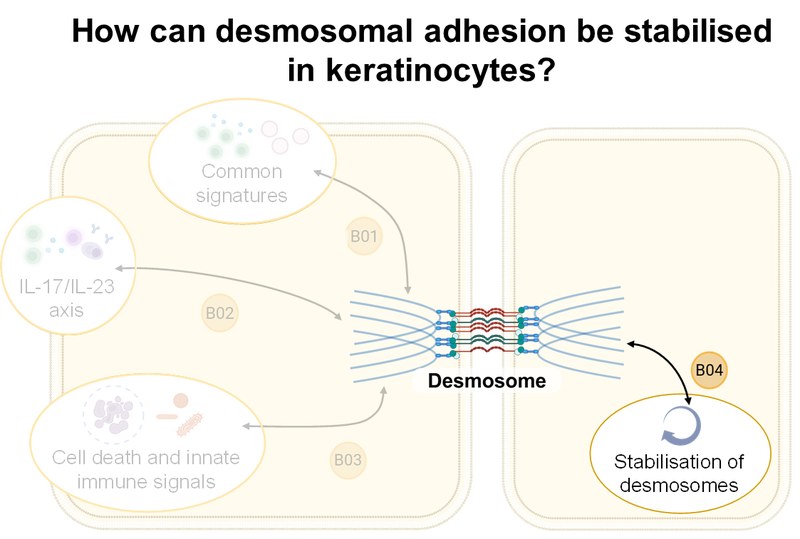
Jens Waschke, Department of Anatomy, LMU Munich
Project: B04 will characterize the composition of desmosomes in desmoglein (Dsg)1- and Dsg3-deficient, as well as Pg-S665 phosphodeficient mice, with respect to desmosome and TJ ultrastructure using STED microscopy and TEM. The phosphodiesterase-4 inhibitor apremilast, the EGFR inhibitor erlotinib, and the Dsg-specific tandem peptide will be used to stabilize keratinocyte adhesion and improve epidermal barrier function. Using multiplex kinome analysis, we will investigate the autoantibody-induced signaling response in detail and verify signaling pathways.
Open Position available! (check for link)
Inhalt ausklappen Inhalt einklappen B05 - Hyper-adhesion and mechanical stress to treat pemphigus
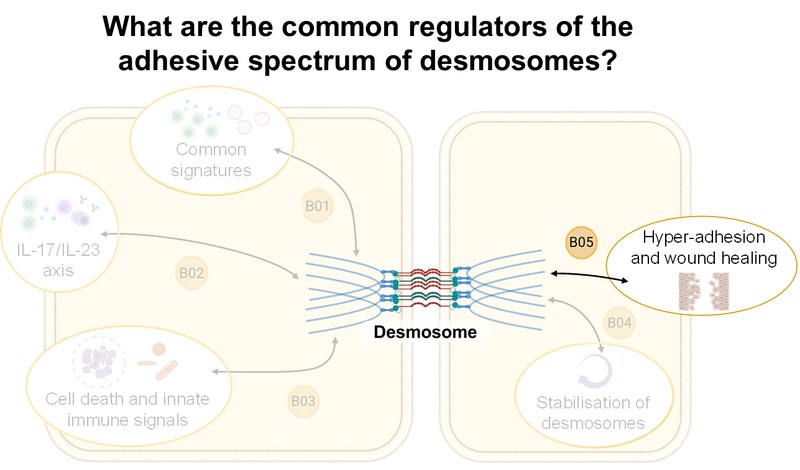
Franziska Hartig-Vielmuth, Department of Anatomy, LMU Munich
Project: B05 will combine biochemical analyses with adhesion analyses and single-molecule force spectroscopy to determine the binding properties of desmosomal cadherins under specific adhesion conditions. Kinome and transcriptome analyses, as well as biochemical approaches, will identify common regulators of the adhesion spectrum and intercellular adhesion in pemphigus.
Open Position available! (check for link)
Inhalt ausklappen Inhalt einklappen B06 - Identification of new treatment options to stabilise cell adhesion in desmosome diseases
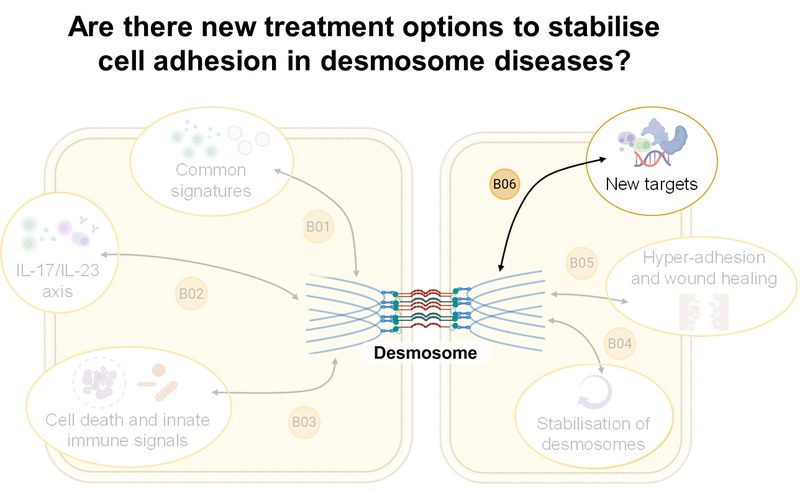
Ralf Ludwig, Institute of Experimental Dermatology, University of Lübeck
Jens Waschke, Department of Anatomy, LMU Munich
Enno Schmidt, Institute of Experimental Dermatology, University of Lübeck
Project: B06 will expand our understanding of autoantibody-induced signaling in keratinocytes in pemphigus and investigate corresponding skin samples. We will then characterize kinome signatures in model systems of the digestive tract, including organoids, as well as patients with IBD and EoE. Further therapeutic interventions with apremilast, sildenafil, erlotinib, midostaurin, and a desmoglein-crosslinking tandem peptide will be tested in preclinical disease models.






