04.10.2021 Following exit from mitosis, RNA pol II is required for reestablishing 3D genome folding by loading cohesin complexes onto DNA
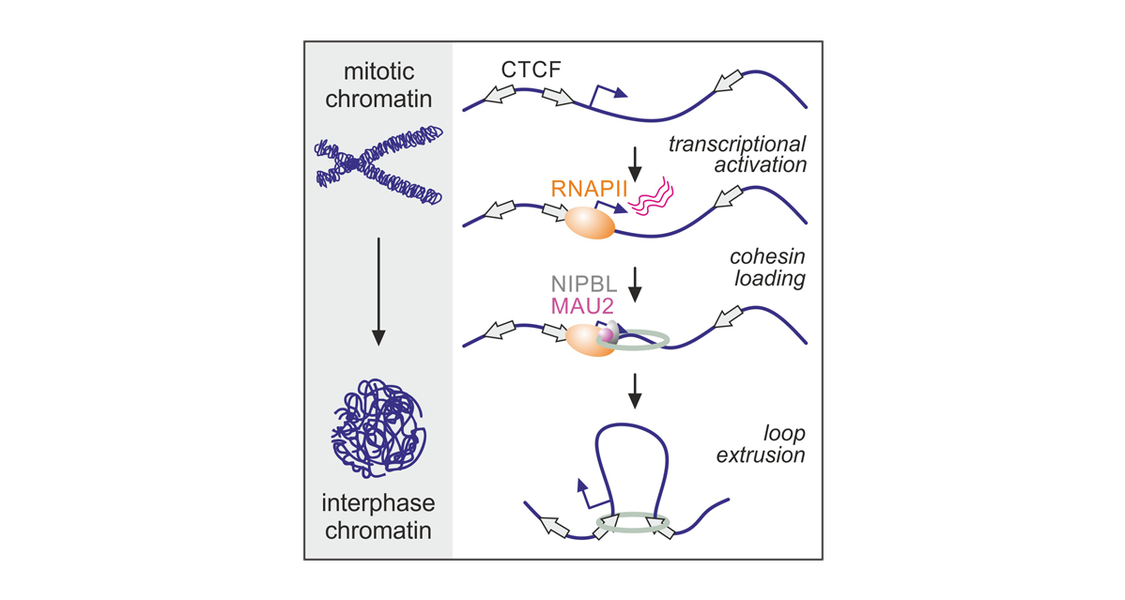
Mammalian chromosomes are intricate three-dimensional(3D) entities, and their 3D structure is intimately linked to the execution of their functions. This 3D structure though changes with every cell division as the process of mitosis induces drastic chromosome condensation. Subsequent reentry into the G1 phase of the cell cycle requires chromosomes to reestablish their 3D interphase organization. We tested the role of RNAPII in this transition using cell that allow for its controllable and acute degradation. Using In situ Hi-C, we were able to show that RNAPII is required for both compartment and loop establishment following mitosis. Evidence from cohesin/CTCF chromatin binding, super-resolution imaging, and in silico modeling pointed to these effects being a result of RNAPII-mediated cohesin loading upon G1 reentry. Our findings reconcile the role of RNAPII in gene expression with that in 3D chromatin architecture.
Full Text Link:Zhang, S., Übelmesser, N., Josipovic, N., Forte, G., Slotman, J.A., Chiang, M., Gothe, H.J., Gusmao, E.G., Becker, C., Altmüller, J., et al. (2021). RNA polymerase II is required for spatial chromatin reorganization following exit from mitosis. Sci Adv 7, eabg8205. 10.1126/sciadv.abg8205.
Contact:
Nadine Übelmesser (PhD Student Göttingen)
Argyris Papantonis (PI Göttingen)
Kerstin Wendt (PI Rotterdam)