Main Content
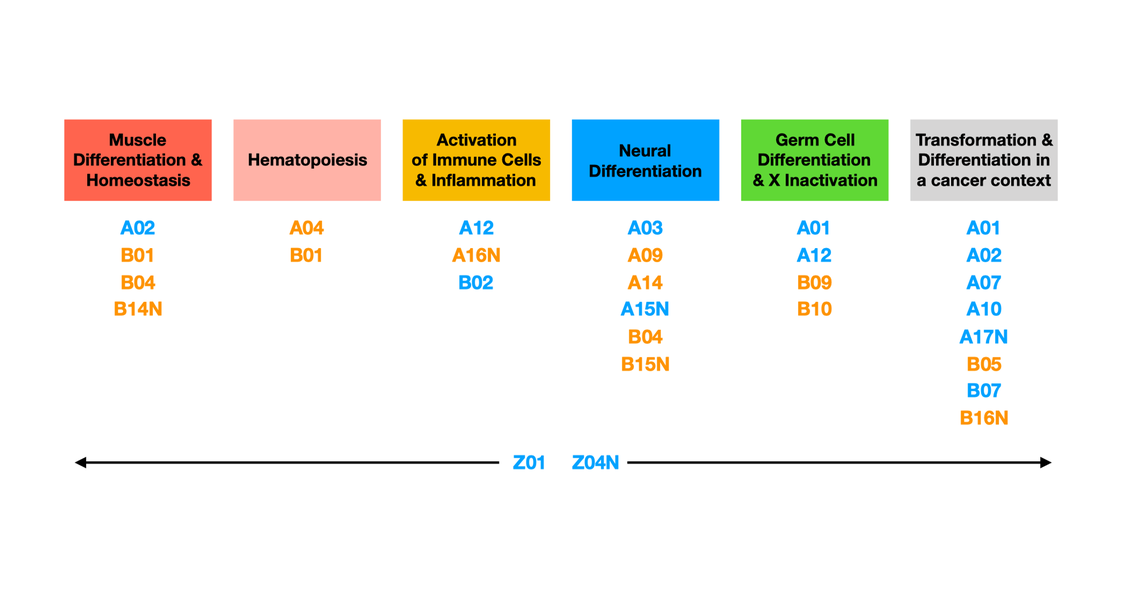
Projects of the TRR 81
The differentiation of cells, in both normal and disease settings, is largely governed by changes to chromatin structure. These changes, which manifest at several levels, determine gene activity and thereby the developmental program in time and space. At a more local level, genome regions containing one or several genes are embedded in a relatively open and accessible chromatin structure and are enriched for active histone marks. Others have a more closed chromatin conformation and predominantly carry repressive histone marks. Changes between these two chromatin states are largely modulated and shaped by DNA binding transcription factors targeting enzymatic functions, such as histone modifying and nucleosome remodelling enzymes, to chromatin. At a more global level, structural proteins such as CTCF or cohesin drive chromatin folding, loop formation, intra- and inter-chromosomal contacts to regulate accessibility and activity of larger genomic regions.
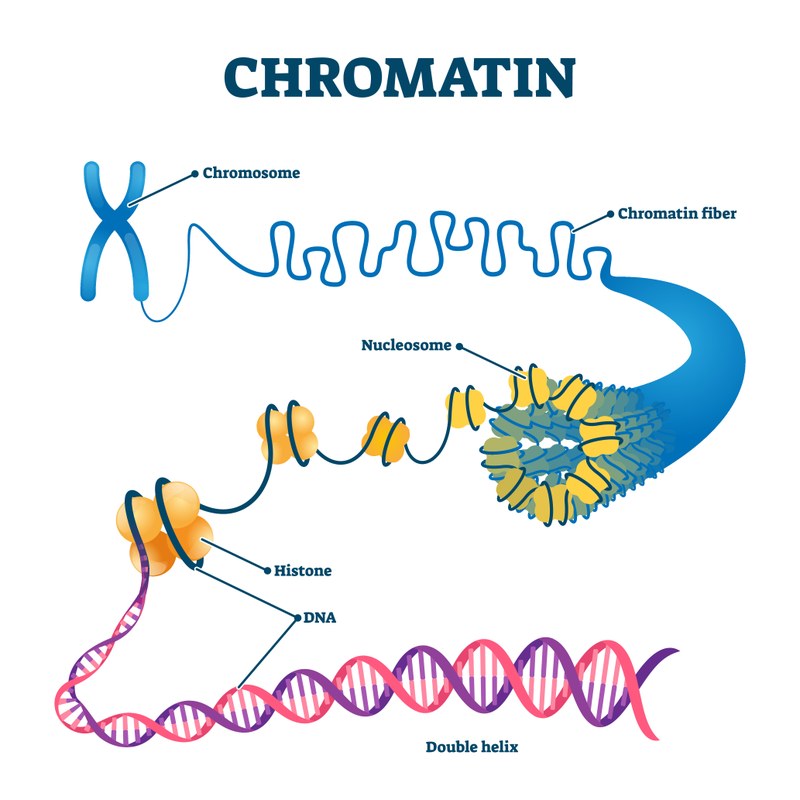
These two modes of chromatin regulation (local and global) are tightly interconnected but serve as a conceptual framework to divide the projects of the TRR81 into two groups: "A projects" focus on transcription factor and enzyme-driven chromatin changes and "B projects" study the genomic organisation in nuclear space.