Main Content
Project Area A - Chromatin changes mediated by enzymatic modification
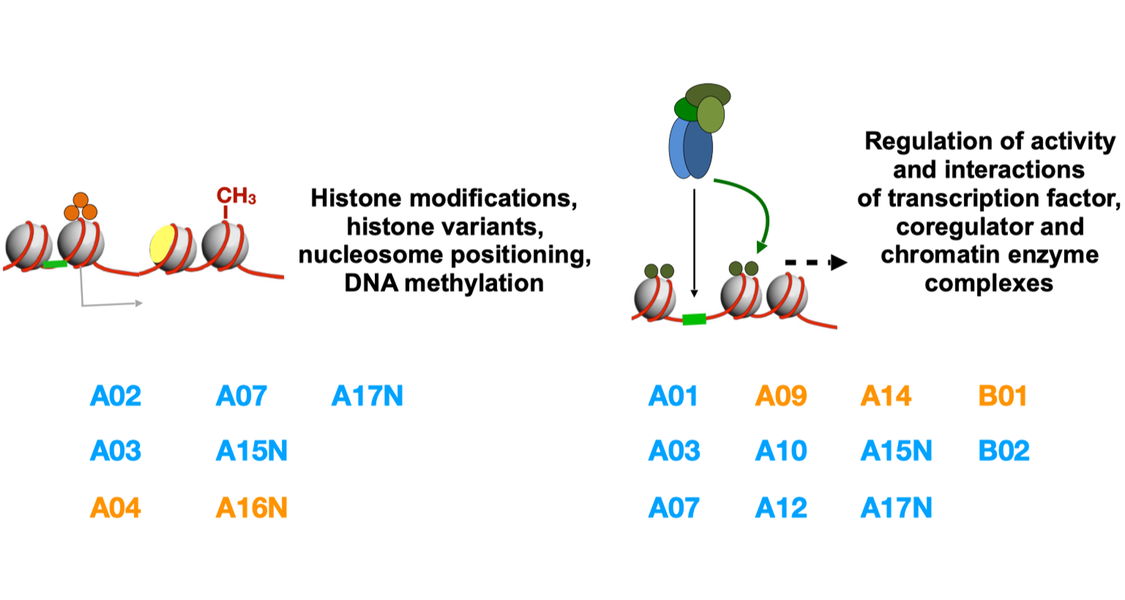
Epigenome regulation by ATP-dependent nucleosome remodelers
Delineating the interplay of epigenetic modifiers and transcription factors in the control of muscle stem cell homeostasis and higher order chromatin organization
Functional analysis of the histone arginine methyltransferase PRMT6
Repurposing CRISPR/Cas9 for targeted demethylation of the gamma-globin promoters
Functional characterization of novel histone phosphorylations
Autism-relevant transcriptional networks in human neural stem cells
Role of p73-directed chromatin alterations for neuroendocrine differentiation in small cell lung cancer
Chromatin regulation mediated by the RBP-J/SHARP corepressor complex
Zeb2 in cell determination regulated by the Activin-Nodal/BMP system: genome-wide binding sites, endogenous partners and long-range locus control
Role of mammalian histone variants H3.3 and H2A.Z in differentiation and disease
Functional epigenomics of type-2 immune responses in allergic asthma
Molecular mechanisms and (patho)physiological consequences of PRC2.1-mediated gene regulation