Main Content
X-Ray crytallographic studies of ligand-binding proteins for compatible solutes and enzymes involved in compatible solute synthesis
X-Ray crystallography provides an atomic view of proteins with unprecedented resolution. To gain deeper inside into the binding of compatible solutes by components of ABC-and TRAP-type transport systems, we carry out in collaboration with laboratories versed in crystallography, the structural analysis of ligand-binding proteins from various types of transport systems. Furthermore, we also analyse the structure of enzymes involved in compatible solute synthesis. Studies are also in progress to obtain the crystal structure of regulatory proteins controlling the expression of genes encoding transport systems for compatible solutes and biosynthetic enzymes. To complement these structural studies of proteins, we carry out biochemical, mutational and functional experiments. We collaborate with the following laboratories:
● Prof. Dr. Wolfram Welte / Prof. Dr. Kay Diederichs (University of Konstanz; Germany)
● Prof. Dr. Lutz Schmitt (University of Düsseldorf; Germany)
● Prof. Dr. Klaus Reuter (University of Marburg; Germany)
● Prof. Dr. Lars Oliver Essen (University of Marburg; Germany)
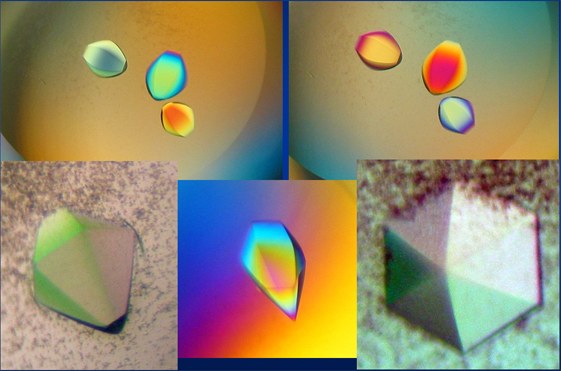
Crystals of the ectoine hydroxylase EctD from Salibacillus salexigens
(Picture provided by Dr. K. Reuter; University of Marburg)
The structural data of the proteins determined in these collaborations have been deposited in the PDB (Protein Data Bank). PDB files can be downloaded from this databank and viewed with PyMol that is available free of charge for educational users running on different operating systems.
● Schiefner, A., J. Breed, L. Bösser, S. Kneip, J. Gade, G. Holtmann, K. Diedrichs, W. Welte and E. Bremer. (2004). Cation-πinteractions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J. Biol. Chem. 279:5588-5596
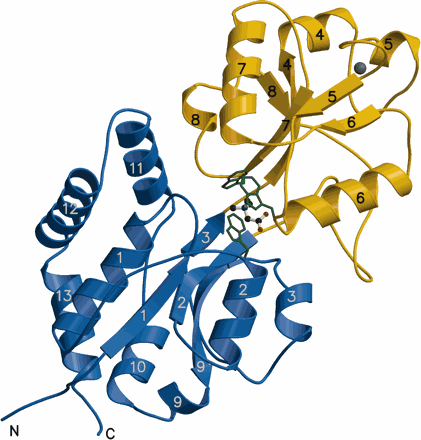
ProX in complex with glycine betaine (PDB Code)
ProX in complex with proline betaine (PDB Code)
- Schiefner, A., G. Holtmann, K. Diederichs, W. Welte and E. Bremer. (2004). Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Biol. Chem. 279:48270-48281.
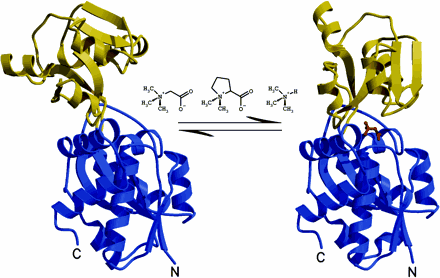
ProX in complex with glycine betaine (PDB Code)
ProX in complex with proline betaine (PDB Code)
ProX without a ligand (PDB Code)
- Horn, C., L. Sohn-Bösser, J. Breed, W. Welte, L. Schmitt and E. Bremer. (2006). Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J. Mol. Biol. 357:592-606.
OpuAC in complex with glycine betaine (PDB Code)
OpuAC in complex with proline betaine (PDB Code)
- Hanekop, N., M. Hoeing, L. Sohn-Bösser, M. Jebbar, L. Schmitt and E. Bremer (2007). Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition for the compatible solutes ectoine and hydroxyectoine. J. Mol. Biol. 374:1237-1250.
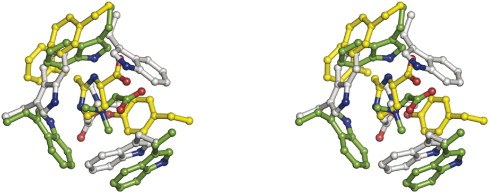
EhuB in complex with ectoine (PDB Code)
EhuB in complex with hydroxyectoine (PDB Code)
● Smits, S. H. J., Höing, M., Lecher, J., Jebbar, M., Schmitt L., and Bremer E. (2008). The compatible solute-binding protein OpuAC from Bacillus subtilis: ligand-binding, site directed mutagenesis and crystallographic studies. J.Bacteriol. 190:5663-5671.
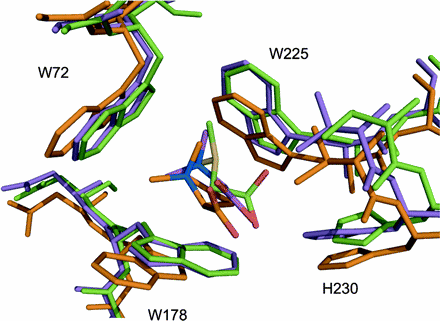
OpuAC in complex with DMSA (PDB Code)
● Oswald, C., Smits, S.H.J., Höing, M., Sohn-Bösser, L., Dupont, L., Le Rudulier, D., Schmitt, L. and Bremer, E. (2008). Crystal structures of the choline/acetylcholine substrate binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded closed states. J. Biol. Chem. 283:32848-32859.

ChoX in complex with choline (PDB Code)
ChoX in complex with acetylcholine (PDB Code)
ChoX in a closed structure without a ligand (PDB Code)
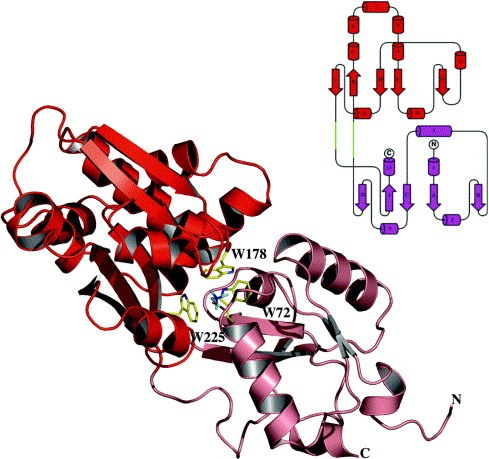
● Lecher, J., Pittelkow, M., Zobel, S., Bursy, J., Bonig, T., Smits, S. H., Schmitt, L. & Bremer, E. (2009). The crystal structure of UehA in complex with ectoine-A comparison with other TRAP-T binding proteins. J. Mol. Biol. 389:58-73.
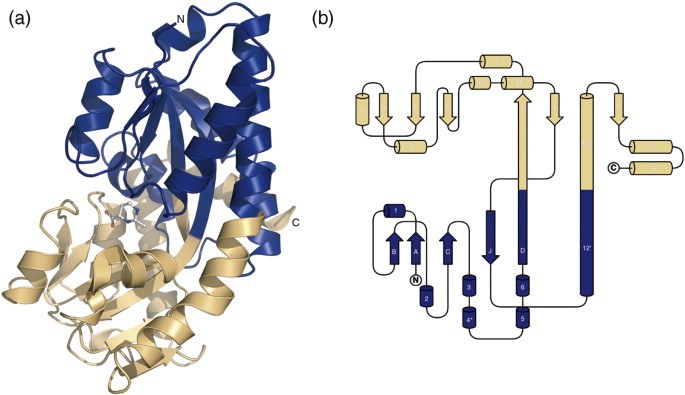
Structure of the UehA binding protein in complex with ectoine (PDB Code)
(a) Overall structure of UehA with the bound ectoine
(b) Topological organization of UehA
● Oswald C, Smits SH, Höing M, Bremer E, Schmitt L (2009). Structural analysis of the choline-binding protein ChoX in a semi-closed and ligand-free conformation. Biol Chem. 390:1163-1170.
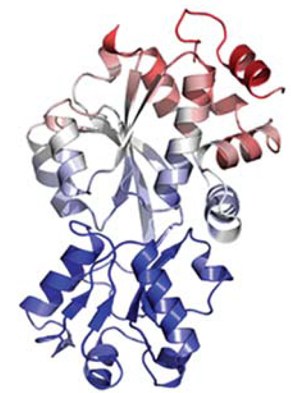
Structure of the choline binding protein ChoX from Sinorhizobium melilotti in a closed but ligand-free conformation (PDB Code)