Main Content
Barrett ’s esophagus / carcinoma
The aim of the research network g4b (genes for barrett’s) is to elucidate the genetic and cell biological causes of Barrett's esophagus and -carcinoma. The molecular genetic analyses for g4b are carried out at the Center for Human Genetics at the University Hospital of Marburg, and at the Institute for Human Genetics at the University Hospital in Bonn. The g4b website can be accessed under www.barrett-konsortium.de.
Clinical features of Barrett's esophagus and carcinoma
Barrett's esophagus usually arises as a result of gastroesophageal reflux, whereby a retrograde flow of gastric acid leads to inflammation of the lower esophagus. Around 10% of patients with gastroesophageal reflux develop Barrett's esophagus (Böhmer et al. (2017) Neurogastroenterol Motil). In this disorder, which is named after the British surgeon Norman Barrett (1903-1979), characteristic changes are observed in the mucous membrane of the esophagus. Symptoms include heartburn and chest pain. A diagnosis of Barrett's esophagus can only be assigned via visualization of the esophagus (endoscopy). To ensure a definitive diagnosis, a tissue sample (biopsy) is also taken.
Barrett's carcinoma is an esophageal cancer that arises secondary to Barrett’s esophagus, in which degenerate cells (cancer cells) are present in the esophagus. Only a small proportion of Barrett's esophagus patients develop Barrett's carcinoma (Böhmer et al. (2017) Neurogastroenterol Motil). In recent years, however, the prevalence of Barrett's carcinoma in developed countries has increased exponentially. Although this is largely attributable to environmental factors, genetic factors are also implicated in the development of the disorder. The symptoms of Barrett's carcinoma often only appear at an advanced stage of disease. The first symptoms are difficulty swallowing, retrosternal pain, and weight loss. Advanced Barrett's carcinoma has a poor prognosis. As with Barrett's esophagus, diagnosis is usually made via endoscopy and tissue biopsy. Confirmation of the presence of a Barrett’s carcinoma requires microscopic examination of the biopsy samples by an experienced pathologist.
Genetics of Barrett's esophagus and -carcinoma
Barrett's esophagus and -carcinoma are multifactorial diseases (Böhmer et al. (2017) Neurogastroenterol Motil). To identify genetic risk variants for Barrett's esophagus and Barrett's carcinoma, we conducted a GWAS in collaboration with research groups from Australia, the UK, and the USA using a large case-control cohort of European origin (> 10,000 patients,> 17,000 controls) (Gharahkhani et al. (2016) Lancet Oncol). A total of > 11 million common genetic variants were tested for association with the disease. In total, the analyses identified 13 genetic risk variants (P <5 x 10-08, odds ratio (OR) = 1.11-1.23) for Barrett's esophagus and -carcinoma. The Manhattan plot in Figure 1 shows the associated risk variants, their chromosomal locations, and genes located in close proximity.
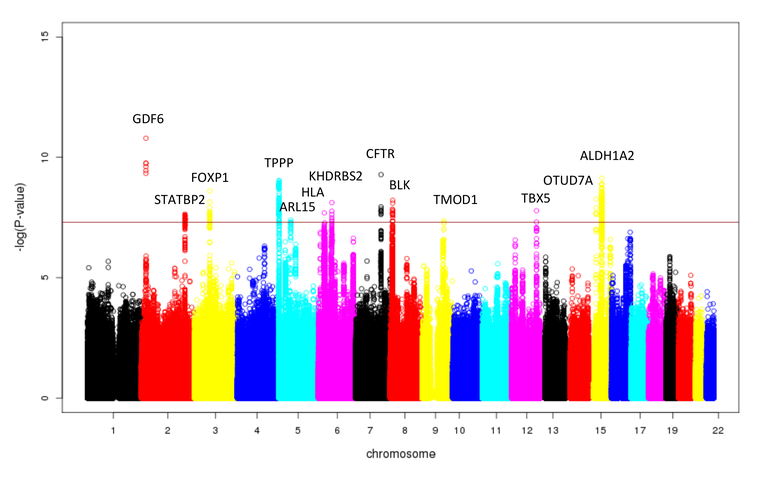
Figure 1: Manhattan plot of the GWAS for Barrett's esophagus and -carcinoma in> 10,000 patients and 17,000 controls. Each point corresponds to a genetic variant. The x-axis shows the chromosomal position of the variants, and the y-axis shows the strength of their association to the disease. A total of 13 variants showed a genome-wide significant association (P <5 x 10-08). Genes in close proximity to the associated variants are indicated above the GWAS signals.
All significantly associated risk variants represent the basis, so to speak, for the development of Barrett's esophagus and -carcinoma. An exception to this is the genetic variant rs9823696 on chromosome 3q27, which is located near the genes HTR3C and ABCC5. This only showed an association in the GWAS of Barrett's carcinoma (P = 1.6 x 10-08, OR = 1.17). In contrast, it was not associated with Barrett's esophagus (P = 0.45, OR = 1.02) (see Figure 2). Such cancer-specific risk variants will be of key importance, since they will enable a prediction of the likelihood that patients with Barrett's esophagus will develop carcinoma.
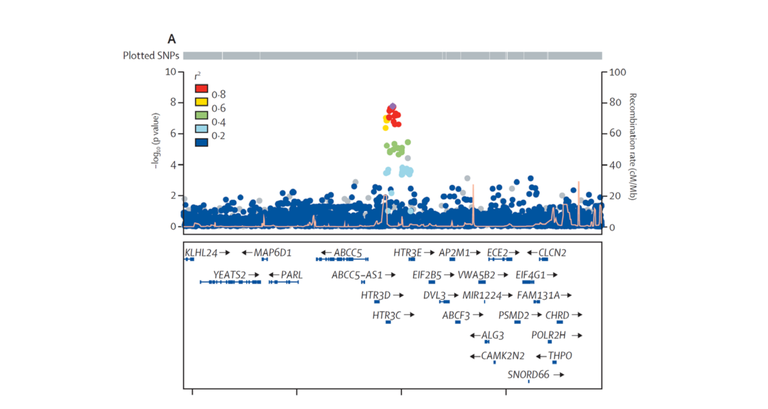
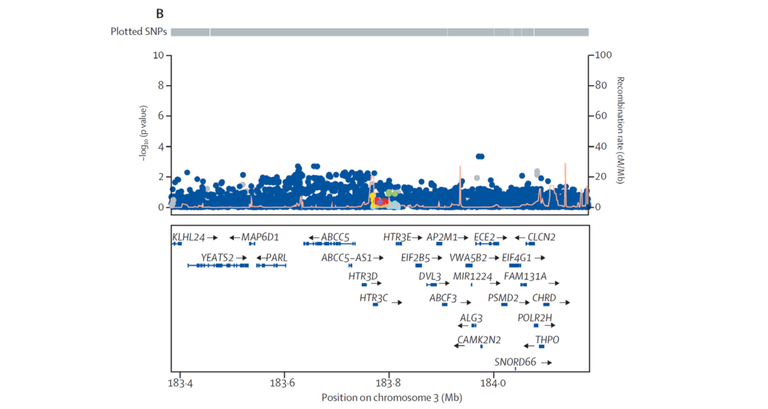
Figure 2: Regional association plot for the Barrett’s carcinoma-specific association near the genes HTR3C and ABCC5. Each point corresponds to a genetic variant. The x-axis shows the location of each variant on chromosome 3q27, and the y-axis shows the strength of their association to the disease. The upper plot shows the genome-wide significant association with Barrett's carcinoma (P <5 x 10-08). The lower plot shows the same region, with no association with Barrett's esophagus.
The GWAS data were also inputted to pathway analyses. These revealed that genes in the proximity of association signals for cellular processes of muscle cell differentiation and mesenchymal cell differentiation were enriched. A muscle differentiation disorder could result in the formation of a diaphragmatic hernia, which is a major predisposing factor for the reflux of gastric acid into the esophagus. In contrast, mesenchymal cell differentiation is an important pathway in terms of the loss of cell adhesion, increased cell migration, and cell invasion. These are important processes in the development of Barrett's carcinoma (Gharahkhani et al. (2016) Lancet Oncol).
The research group is currently engaged in the functional characterization of the identified risk variants. In addition, investigations are being conducted to determine a common genetic influence for Barrett's esophagus / carcinoma and other phenotypes, e.g., obesity, as determined at the level of polygenic risk scores (PRSs), while the patient cohort is being enlarged for the purposes of an extended GWAS.
Contact person
Prof. Dr. Johannes Schumacher
The g4b webpage can be accessed under www.barrett-konsortium.de.
Selected Publications
Schröder J, Schüller V, May A, Gerges C, Anders M, Becker J, Hess T, Kreuser N, Thieme R, Ludwig KU, Noder T, Venerito M, Veits L, Schmidt T, Fuchs C, Izbicki JR, Hölscher AH, Dakkak D, Jansen-Winkeln B, Moulla Y, Lyros O, Niebisch S, Mehdorn M, Lang H, Lorenz D, Schumacher B, Mayershofer R, Vashist Y, Ott K, Vieth M, Weismüller J, Mangold E, Nöthen MM, Moebus S, Knapp M, Neuhaus H, Rösch T, Ell C, Gockel I, Schumacher J, Böhmer AC. Identification of loci of functional relevance to Barrett's esophagus and esophageal adenocarcinoma: Cross- referencing of expression quantitative trait loci data from disease-relevant tissues with genetic association data. PLoS One. 2019 Dec 31;14(12):e0227072.
An J, Gharahkhani P, Law MH, Ong JS, Han X, Olsen CM, Neale RE, Lai J, Vaughan TL, Böhmer AC, Jankowski J, Fitzgerald RC, Schumacher J, Palles C, BEACON, 23andMe Research Team, Whiteman DC, MacGregor S. Gastroesophageal reflux GWAS identifies risk loci that also associate with subsequent severe esophageal diseases. Nat Commun. 2019 Sep 16;10(1):4219.
Böhmer AC, Schumacher J. Insights into the genetics of gastroesophageal reflux disease (GERD) and GERD-related disorders. Neurogastroenterol Motil 2017 29.
Gharahkhani P, Fitzgerald RC, Vaughan TL, Palles C, Gockel I, Tomlinson I, Buas MF, May A, Gerges C, Anders M, Becker J, Kreuser N, Noder T, Venerito M, Veits L, Schmidt T, Manner H, Schmidt C, Hess T, Böhmer AC, Izbicki JR, Hölscher AH, Lang H, Lorenz D, Schumacher B, Hackelsberger A, Mayershofer R, Pech O, Vashist Y, Ott K, Vieth M, Weismüller J, Nöthen MM; Barrett's and Esophageal Adenocarcinoma Consortium (BEACON); Esophageal Adenocarcinoma GenEtics Consortium (EAGLE); Wellcome Trust Case Control Consortium 2 (WTCCC2), Attwood S, Barr H, Chegwidden L, de Caestecker J, Harrison R, Love SB, MacDonald D, Moayyedi P, Prenen H, Watson RGP, Iyer PG, Anderson LA, Bernstein L, Chow WH, Hardie LJ, Lagergren J, Liu G, Risch HA, Wu AH, Ye W, Bird NC, Shaheen NJ, Gammon MD, Corley DA, Caldas C, Moebus S, Knapp M, Peters WHM, Neuhaus H, Rösch T, Ell C, MacGregor S, Pharoah P, Whiteman DC, Jankowski J, Schumacher J. Genome-wide association studies in oesophageal adenocarcinoma and Barrett's oesophagus: a large-scale meta-analysis. Lancet Oncol 2016 17: 1363-73.
Becker J, May A, Gerges C, Anders M, Schmidt C, Veits L, Noder T, Mayershofer R, Kreuser N, Manner H, Venerito M, Hofer JH, Lyros O, Ahlbrand CJ, Arras M, Hofer S, Heinrichs SK, Weise K, Hess T, Böhmer AC, Kosiol N, Kiesslich R, Izbicki JR, Hölscher AH, Bollschweiler E, Malfertheiner P, Lang H, Moehler M, Lorenz D, Ott K, Schmidt T, Nöthen MM, Hackelsberger A, Schumacher B, Pech O, Vashist Y, Vieth M, Weismüller J, Knapp M, Neuhaus H, Rösch T, Ell C, Gockel I, Schumacher J. The Barrett-associated variants at GDF7 and TBX5 also increase esophageal adenocarcinoma risk. Cancer Med 2016 5: 888-91.
Becker J, May A, Gerges C, Anders M, Veits L, Weise K, Czamara D, Lyros O, Manner H, Terheggen G, Venerito M, Noder T, Mayershofer R, Hofer JH, Karch HW, Ahlbrand CJ, Arras M, Hofer S, Mangold E, Heilmann-Heimbach S, Heinrichs SK, Hess T, Kiesslich R, Izbicki JR, Hölscher AH, Bollschweiler E, Malfertheiner P, Lang H, Moehler M, Lorenz D, Müller-Myhsok B, Ott K, Schmidt T, Whiteman DC, Vaughan TL, Nöthen MM, Hackelsberger A, Schumacher B, Pech O, Vashist Y, Vieth M, Weismüller J, Neuhaus H, Rösch T, Ell C, Gockel I, Schumacher J. Supportive evidence for FOXP1, BARX1, and FOXF1 as genetic risk loci for the development of esophageal adenocarcinoma. Cancer Med 2015 4: 1700-4.