Main Content
Expression quantitative trait locus (eQTL) analyses
Modern technologies have enabled the integrative evaluation of the genome (DNA) and the transcriptome (mRNA). This in turn has allowed the development of expression quantitative trait locus (eQTL) analyses, in which the alleles or genotypes of genetic variants are correlated with the (quantitative) level of transcript expression. For the first time, this approach has enabled the identification of genetic variants that regulate the level of gene expression (see Figure 1).
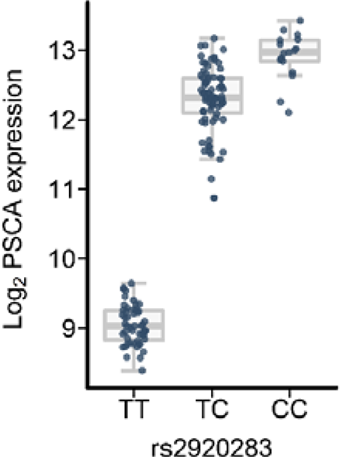
Figure 1: To exemplify this concept, the expression level of the gene PSCA is shown as a function of the genotype distribution of variant rs2223286. The y-axis shows the PSCA transcript level in the gastric mucosa. Each point corresponds to one subject. The subjects are classified according to rs2223286 genotype status (x-axis). The box plot shows that allele C leads to an induction of PSCA expression in the gastric mucosa, whereas allele T leads to a reduction in PSCA expression (Heinrichs et al. (2018) Cancer Med). In a previous genome-wide association study (GWAS), the C allele of this variant was identified as a risk factor for gastric carcinoma (Abnet et al. (2010) Nat Genet). The GWAS risk variant was functionally characterized using the depicted eQTL analysis.
For several years, eQTL analyses have also been possible across genomes and transcripts, and this development has enabled the systematic ascertainment of gene regulation processes. Using maximum genomic coverage, genetic variants are examined, and their genotype distributions are then correlated with the expression level of all ascertainable transcripts.
A highly innovative eQTL approach is to record gene regulatory processes after exogenous cell stimulation (Kim-Hellmuth et al. (2017) Nat Commun, Kim et al. (2014) Nat Commun). This approach takes into account the fact that the influence of genetic variability on gene expression often only becomes apparent in the context of activated cell biological processes, i.e., after exogenous stimulation or the influence of environmental factors (see Figure 2).
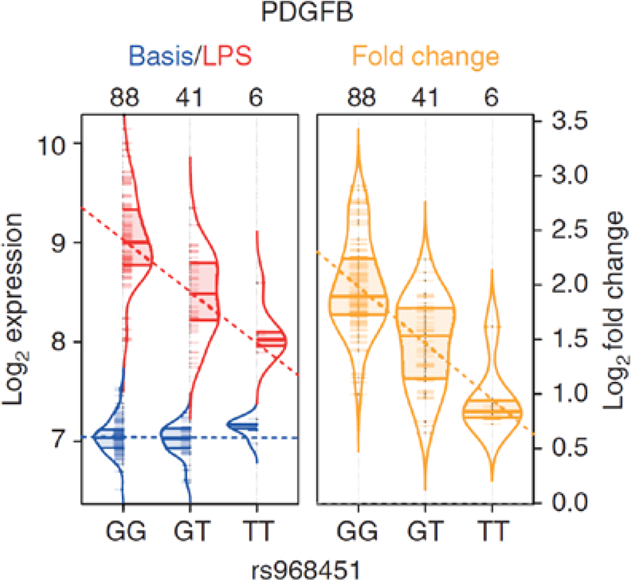
Figure 2: To exemplify this concept, an eQTL in monocytes that is only apparent after stimulation with lipopolysaccharides (LPS) is shown. LPS is a component of the bacterial outer membrane and triggers an immune response in monocytes by binding to Toll-like-4 receptors (TLR4). The y-axis shows the transcript level of the gene PDGFB, and the x-axis shows the genotype of variant rs968451. Blue indicates the absence of any eQTL effect in unstimulated monocytes. Red indicates the induction of PDGFB expression in monocytes that have been stimulated by LPS. This demonstrates an eQTL effect in which a stronger induction of expression occurs in carriers of allele G. The right violin plot (yellow) shows PDGFB expression before and after LPS stimulation in relation to genotype distribution (cited from Kim et al. (2014) Nat Commun).
Stimulus-specific or exposure-eQTLs (e2QTLs) are of major scientific importance. They occur at the very beginning of an activated cell biological process, and are thus of key relevance in terms of inter-individual differences in the reaction of the organism to a stimulus. In accordance with this, a separate eQTL analysis of monocytes that were stimulated with bacterial and viral components, and non-stimulated monocytes, showed that 3-18% of all eQTLs were stimulus-specific (Kim-Hellmuth et al. (2017) Nat Commun).
In principle, eQTL analyses are not restricted to the detection of regulated genes and the associated variants. They also enable the identification of those cell biological networks or signaling pathways downstream of the stimulus with which the tissue reacts to exogenous influences (see Figure 3).
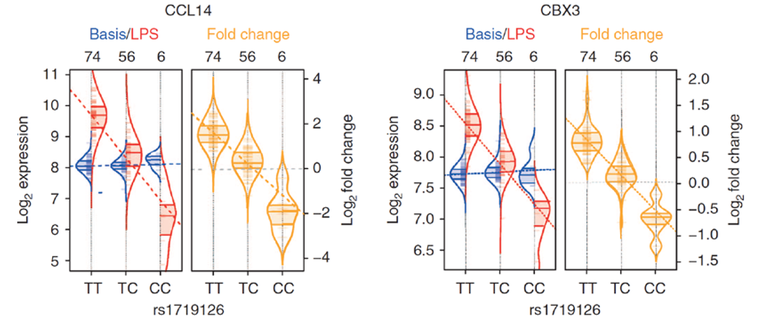
Figure 3: To exemplify this concept, two eQTLs that are only active in LPS-stimulated monocytes are shown (for explanation, see Figure 2). The right violin plot shows a cis-eQTL between SNP rs1719126 and the gene CCL14, which lie in close proximity on chromosome 17. The left violin plot shows a trans-eQTL between the same SNP and the CBX3 gene, which is located on chromosome 7. CCL14 encodes a chemokine, and displays strong allele-specific expression differences in the context of the LPS-induced immune response. An almost identical eQTL effect is found for the expression of CBX3, a transcription factor in the context of the immune response (Smallwood et al. (2012) Genome Research). The eQTL analysis suggests that rs1719126 regulates the expression of CBX3 via the “mediator transcript” CCL14, a connection that was hitherto completely unknown (cited from Kim et al. (2014) Nat Commun).
The eQTL analysis approach is also of major importance in terms of the functional processing of risk variants for multifactorial disease that have been identified via genome-wide association studies (GWAS). For example, rs968451 from Figure 2 was identified as a risk variant in a GWAS of primary biliary cirrhosis (PBC) (Mells et al. (2011) Nat Genet). Due to its location in an intergenic region, the research group was unable to ascertain either the disease-relevant gene or the underlying pathomechanism. Even with the then available eQTL data on non-activated cells, the researchers could not prioritize any PBC disease gene in this region on which rs968451 exerts an influence. The eQTL analysis of LPS-stimulated monocytes (Kim et al. (2014) Nat Commun) now suggests that PDGFB is the PBC-relevant disease gene, and that reduced expression (allele T was the risk allele in GWAS) secondary to TLR4- Activation is the pathomechanism.
In summary, the outlined eQTL concept enables the identification of regulatory variants and associated genes, as well as signaling pathways that are only active after the induction of a cell biological process via exogenous stimulation. They are of major clinical importance, since the pathophysiology of many diseases only manifests via interactions with exogenous influencing factors. Stimulation-specific eQTLs occur at the beginning of the pathophysiologically-relevant process, and determine inter-individual differences in phenotypic characteristics.
In collaboration with other research teams, our group is currently working on e2QTL analyses of diverse metabolic, inflammatory, cardiovascular, oncological, and infectious diseases.
Selected Publications
Kim-Hellmuth S, Bechheim M, Pütz B, Mohammadi P, Nédélec Y, Giangreco N, Becker J, Kaiser V, Fricker N, Beier E, Boor P, Castel SE, Nöthen MM, Barreiro LB, Pickrell JK, Müller-Myhsok B, Lappalainen T, Schumacher J, Hornung V. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun 2017 8: 266.
Kim S, Becker J, Bechheim M, Kaiser V, Noursadeghi M, Fricker N, Beier E, Klaschik S, Boor P, Hess T, Hofmann A, Holdenrieder S, Wendland JR, Fröhlich H, Hartmann G, Nöthen MM, Müller-Myhsok B, Pütz B, Hornung V, Schumacher J. Characterizing the genetic basis of innate immune response in TLR4-activated human monocytes. Nat Commun 2014 5: 5236.
Becker J, Wendland JR, Haenisch B, Nöthen MM, Schumacher J. A systematic eQTL study of cis-trans epistasis in 210 HapMap individuals. Eur J Hum Genet 2012 20: 97-101.