Main Content
Methods
Our institute employs a wide range of state-of-the-art molecular methods for the comprehensive analysis of tumors.
Using next-generation sequencing (NGS), we analyze more than 400 relevant genes and detect genetic alterations such as point mutations, gene fusions, and copy number variations. This is complemented by whole-exome sequencing, which covers the entire protein-coding genome, as well as liquid biopsy panel analyses that minimally invasively detect circulating tumor DNA in the blood and enable real-time monitoring of tumor dynamics.
For spatial analysis of gene and protein expression, we use advanced technologies such as 10x Visium, CellDIVE, and GeoMX. These methods provide insights into the tumor microenvironment, immune cell populations, and intratumoral heterogeneity.
Specialized tests such as EndoPredict for breast cancer, HRD analyses to assess DNA repair deficiencies, and the rapid Idylla PCR test support individualized therapy decision-making. Computer-assisted digital image analysis further complements the molecular data through quantitative evaluation of histological sections.
Next-generation sequencing of gene panels
Our next-generation sequencing (NGS) panels enable comprehensive molecular genetic analysis of over 400 cancer-associated genes. This includes the precise detection of point mutations (SNVs), insertions and deletions (indels), copy number alterations (CNAs), gene fusions, and exon-skipping events. In addition, complex genomic biomarkers such as microsatellite instability (MSI) and tumor mutational burden (TMB) are assessed.
The interpretation of detected variants is carried out using established databases to determine their therapeutic relevance. The results are incorporated into personalized treatment decisions within the framework of the interdisciplinary Molecular Tumor Board.
Liquid Biopsy NGS Panel
The liquid biopsy technique enables minimally invasive molecular characterization of tumors by detecting circulating tumor DNA (ctDNA) in blood plasma. This method allows real-time monitoring of tumor dynamics, early detection of therapy-associated resistance mechanisms, and long-term tracking of molecular changes. The data obtained are analyzed using bioinformatic methods and support the individual adaptation of therapy in ongoing clinical trials as well as routine diagnostics.
Whole Exome Sequenzierung
Whole-exome sequencing (WES) enables comprehensive analysis of all protein-coding genes within the genome. In addition to identifying SNVs, indels, and CNAs, it also detects complex molecular markers such as microsatellite instability (MSI), tumor mutational burden (TMB), and homologous recombination deficiency (HRD). Annotation and interpretation of genetic variants are performed using curated databases to assess their clinical relevance. WES serves as a foundation for precise, personalized therapy decisions and is utilized within the framework of the Molecular Tumor Board.
Gene Expression Profiling (whole transcriptome)
RNA sequencing enables comprehensive quantification of gene expression in solid tumors. The resulting transcriptomic data allow for functional characterization of tumors, molecular subtyping, and the development and validation of prognostic and predictive biomarkers. The results are used for gene set enrichment analyses, multivariate omics integrations, and therapy stratification in clinical studies.
HPV – Human Papillomavirus
Testing for human papillomavirus (HPV) is used for the early detection of infections with high-risk HPV types that are associated with the development of various tumors—particularly cervical cancer. Using a simple swab sample, an infection can be detected at an early stage, allowing for regular monitoring and timely intervention to prevent tumor progression.
HRD – Homologous Recombination Deficiency
The analysis of homologous recombination deficiency (HRD) identifies tumors with impaired ability to repair DNA double-strand breaks, as seen in ovarian or breast cancer. A positive HRD status may indicate sensitivity to PARP inhibitors and thus serves as a critical predictive biomarker for therapy planning.
Endopredict
EndoPredict is a molecular genetic test used for risk stratification in patients with hormone receptor-positive, HER2-negative early-stage breast cancer. The test is based on the analysis of a defined gene expression panel in combination with clinicopathological parameters. Its goal is to provide a precise estimate of the risk of metastasis within ten years, thereby supporting informed decisions regarding the need for adjuvant chemotherapy.
Idylla
Idylla is a fully automated, qPCR-based diagnostic system that enables rapid and standardized analysis of therapy-relevant mutations and biomarkers directly from FFPE tissue samples. Thanks to its closed system, results are generated within a very short time, allowing for immediate decision-making in routine clinical practice.
10x Visum
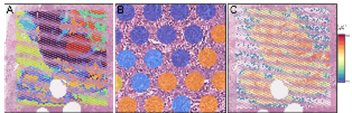
The 10x Visium Spatial Gene Expression Analysis enables precise mapping of gene expression patterns to the histological structure of tissue samples. By combining tissue section imaging with RNA sequencing, this method provides an integrative view of the spatial organization of gene activity within the tumor. This allows for the identification of differential expression patterns in the context of tumor heterogeneity and the microenvironment. The technique offers new insights into the biological processes of tumor development and provides an important basis for the discovery of spatially defined biomarkers.
Genomic profiling using large multi-gene and liquid biopsy panels
By employing large DNA- and RNA-based multigene panels, the institute offers comprehensive molecular profiling of solid tumors. Hundreds of genes representing key oncogenic signaling pathways are analyzed—including mutations, gene fusions, amplifications, and complex biomarkers such as TMB and MSI. Additionally, a newly established liquid biopsy method enables the analysis of circulating tumor DNA from plasma samples, allowing real-time monitoring of molecular changes and early detection of resistance mechanisms.
The interpretation of genomic data is performed using the clinical software solution MH Guide (Molecular Health), which identifies approved therapies, potentially effective agents, and ongoing clinical trials. This platform supports precision oncology decisions within the interdisciplinary tumor board and makes an important contribution to translational research, for example within the framework of the MOMENTUM registry.
Cell Dive – Multiplex imaging technology

CellDIVE is an image-based multiplex immunofluorescence technology that enables simultaneous analysis of up to 60 protein markers in a single tissue section. This allows precise mapping of the composition and activation status of immune cells as well as their interactions with tumor cells. The method is used, among other applications, in translational research projects to characterize the tumor microenvironment.
TMA - Tissue Microarrays
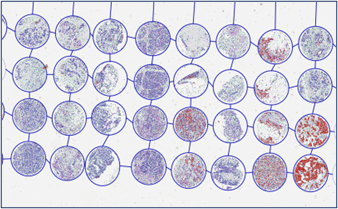
Tissue microarrays (TMAs) enable the parallel analysis of numerous tissue samples under standardized conditions. Small tissue cores from various tumors are combined into a single block, allowing highly efficient studies of biomarker expression in large cohorts. This method is especially used in the preclinical validation of predictive markers.
GeoMX - Digital Spatial Profiling
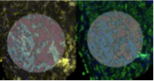
GeoMX is a technology for high-resolution, multiplex analysis of protein and RNA expression directly within the tissue context. It provides detailed information about the spatial distribution of molecular signatures within tumor tissue and its microenvironment. This method helps identify new therapeutically relevant targets and biomarkers in the context of cell interactions.
Digital Image Analysis
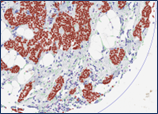
Computer-assisted image analysis enables the quantitative evaluation of histological specimens as well as precise mapping of cellular distributions within the tumor and its microenvironment. In combination with multiplex technologies such as CellDIVE or GeoMX, it facilitates automated detection of functional patterns and supports the identification of new diagnostic and prognostic markers.