Main Content
Regulation of chromatin function by arginine methylation
Posttranslational modifications of histones
Accessibility of the DNA for nuclear processes, such as transcription, replication and repair, is controlled by several mechanisms: DNA methylation, nucleosome remodeling and posttranslational modifications of histones. Histone modifications, including acetylation, phosphorylation and methylation, determine the compaction state and structure of chromatin depending on their nature, localization and combination establishing the so-called "histone code" (Strahl and Allis, 2000). For example di- and trimethylation of histone H3 at lysine 9 (K9) and lysine 27 (K27) predominantly occur in regions of inactive genes and heterochromatin, whereas hyperacetylation of histones and H3 K4 trimethylation are known to mark the promoter regions of actively transcribed genes (Barski et al., 2007). On the mechanistic level, histone modifications regulate chromatin binding of effector proteins that are involved in the generation of different chromatin states. (Jenuwein and Allis, 2001; Turner, 2002). Histone modifications, which are propagated throughout cell division, belong to the epigenetic information. In general, histone-modifying enzymes control fundamental cellular processes, including cell cycle progression and proliferation, differentiation as well as apoptosis. Accordingly, deregulated function or mutation of these enzymes is associated with the onset of cancer and other diseases.
Protein arginine methyltransferases (PRMTs)
Arginine methylation of proteins is catalyzed by a family of enzymes, the PRMTs (protein arginine methyltransferases), which are evolutionary conserved in eukaryotes from fungi to plants and mammals (Di Lorenzo and Bedford, 2011). In the human genome nine PRMT members (PRMT1-9) are encoded. PRMTs share a central highly conserved catalytic domain and perform mono- and dimethylation of the guanidino group of arginine residues using S-adenosylmethionine (SAM) as the methyl donor. With regard to the dimethylation product PRMTs are subdivided into type I enzymes (PRMT1-4, PRMT6 & 8), which catalyze the formation of the asymmetric NG,NG-dimethyl-arginine, and the type II enzyme subfamily (PRMT5 & 7), which generates symmetric NG,NG-dimethylation (Yang and Bedford, 2013). Unmodified and monomethyl arginines can be converted to citrulline by the peptidyl arginine deiminase PAD4 or dimethyl arginines are potentially demethylated (Di Lorenzo and Bedford, 2011). Knockout mice revealed an essential function of PRMT1, PRMT4 and PRMT5 during embryogenesis. Furthermore, deregulated expression of several PRMT members, in particular PRMT1, PRMT4, PRMT5 and PRMT6, is associated with the occurrence of certain tumor types (Yang and Bedford, 2013). PRMTs are implicated in the regulation of a plethora of cellular processes: RNA processing, ribosome biogenesis, nucleo-cytoplasmic transport and chromatin-associated processes, such as DNA-repair, imprinting and transcriptional regulation. Our work concentrates on the chromatin-regulatory functions of arginine methylation and the following projects:
Projects
1. Transcriptional coactivators PRMT1 and PRMT4
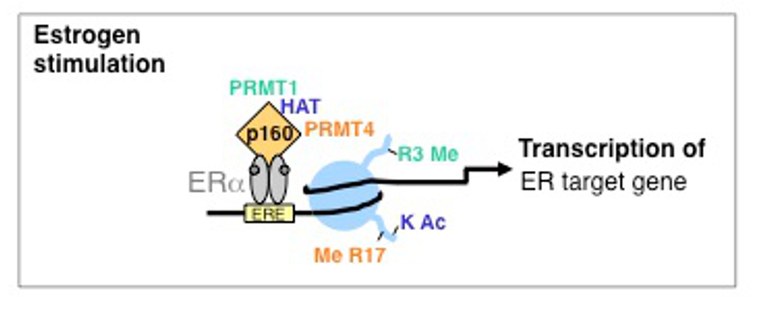
PRMTs are themselves not able to directly interact with DNA. To fulfill their function as transcriptional coregulators the enzymes interact with various transcription factors and thereby are recruited to corresponding target gene promoters. Subsequently, PRMTs affect gene activity in a methyltransferase-dependent manner by modifying histones and other chromatin-proteins. In this project we search for novel gene-regulatory pathways, which might be affected by PRMT1 and PRMT4 and by which mechanism. We found that PRMT4, the first PRMT member that has been linked to transcriptional regulation, methylates arginine 17 (R17) of histone H3 in promoter regions and is an important coactivator of nuclear hormone receptor (NR)-mediated gene expression (Bauer et al., 2002). Furthermore, we identified PRMT1-mediated H4R3 methylation as an early event in NR-induced gene expression, which plays a crucial role in the activation of a subset of estrogen receptor (ER)-target genes (Balint et al., 2005; Wagner et al., 2006). Previous reports suggested that PRMT1 and PRMT4 synergize in their coactivator function and cooperate also with histone acetyltransferases (HATs) (An et al., 2004). Mechanistically, we could show that PRMT1-mediated histone methylation occurs early during activation of NR-target genes and is a prerequisite for promoter hyperacetylation (Wagner et al., 2006). Histone acetylation by CBP/p300 subsequently facilitates promoter recognition by PRMT4 and H3R17 methylation (Daujat et al., 2002). Our work indicates that also other transcription factors are synergistically coactivated by the histone methyltransferase activity of both PRMTs, such as the transcription factor STAT5 (Kleinschmidt et al., 2008).
2. Cooperation between PRMT4 and the chromatin remodeler Mi2 (CHD3/CHD4)
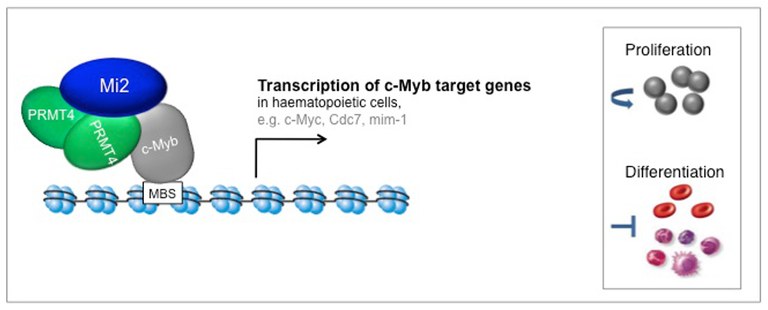
In gel filtration analysis we detected PRMT4 in higher molecular weight complexes than expected from its monomeric or dimeric molecular weight. This suggested that PRMT4 might exert its functions not as an individual protein, but in association with other proteins, similar to what has been described for other chromatin modifiers and transcriptional coregulators. Therefore, we biochemically purified endogenous PRMT4 and identified co-purifying proteins by mass spectrometry analysis. Thus, we discovered the ATP-dependent chromatin remodelers Mi2α and Mi2β, also called CHD3 and CHD4 (chromodomain-helicase-DNA binding protein 3 and 4) respectively, as such candidate interaction partners (Streubel et al., 2013). PRMT4 binds Mi2α and its close relative Mi2β, but not the other components of the repressive Mi2-containing NuRD complex. In search of the biological role of this interaction we found that PRMT4 and Mi2α/β interact with the transcription factor c-Myb and cooperatively coactivate c-Myb target gene expression in haematopoietic cell lines. Depletion of PRMT4 or Mi2α/β in haematopoietic cells resulted in diminished transcriptional induction of c-Myb target genes, attenuated cell growth and survival and in deregulated differentiation resembling the effects caused by c-Myb depletion. These findings reveal an important and so far unknown connection between PRMT4 and the chromatin remodeler Mi2 in c-Myb signaling (Streubel et al., 2013).
3. Transcriptional corepressor PRMT6
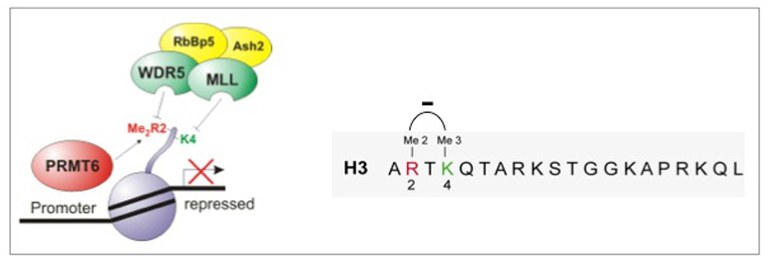
PRMT6 has recently been shown to regulate DNA repair and viral transcription. Given that PRMT6 predominantly resides in the nucleus, we asked whether it possesses methyltransferase activity towards histones and influences cellular gene expression. We found that PRMT6 methylates histone H3 at R2 and histones H4/H2A at R3 in vitro. Overexpression and knockdown of PRMT6 identify the enzyme as the major H3R2 methyltransferase in vivo. H3R2 dimethylation (H3R2me2) is a negative regulator of H3K4 trimethylation (H3K4me3), since it inhibits the H3K4 methyltransferase MLL1 and recruitment of WDR5, a subunit of the MLL K4 methyltransferase complex, to histone H3. By antagonizing the active mark H3K4me3, PRMT6-mediated H3R2me2 results in transcriptional repression of HoxA genes or Myc target genes (Hyllus et al., 2007). In search of the cellular function of PRMT6, our data indicated that the gene-repressive activity of PRMT6 enhances cell proliferation and blocks differentiation as well as senescence, which might explain why overexpression of PRMT6 occurs in several human cancer types and seems beneficial (Stein et al., 2012).
References
An, W. et al. (2004), Cell 117, 735-748.
Balint, B.L. et al. (2005), Molecular and Cellular Biology 25, 5648-5663.
Barski, A. et al. (2007), Cell 129, 823-837.
Bauer, U.M. et al. (2002), EMBO Reports 3, 39-44.
Daujat, S. et al. (2002), Current Biology 12, 2090-2097.
Di Lorenzo, A. & Bedford, M.T. (2011), FEBS Letters 585, 2024-2031.
Hyllus, D. et al. (2007), Genes & Development 21, 3369-3380.
Jenuwein, T. and Allis, D.C. (2001), Science 493, 1074-1080.
Kleinschmidt, M.A. et al., (2008), Nucleic Acids Research 36, 3202-3213.
Strahl, B. and Allis, D.C. (2000), Nature 403, 41-45.
Stein, C. et al. (2012), Nucleic Acids Research 40, 9522-9533.
Streubel, G. et al. (2013), PLoS Genetics 9, e1003343.
Turner, B.M. (2002), Cell 111, 285-291.
Wagner, S. et al. (2006), Journal of Biological Chemistry 281, 27242-27250.
Yang, Y. and Bedford, M.T. (2013), Nature Reviews Cancer 13, 37-50.