Main Content
CHD4/dMi-2 complexes & regulation of gene expression
Jonathan Lenz
CHD4 is an ATP-dependent remodeller that modifies chromatin structure by sliding nucleosomes along the genome. This change in nucleosome density impacts many DNA-related processes like the transcription of genes and DNA repair. To facilitate these functions, CHD4 interacts with other chromatin regulators and transcription factors.
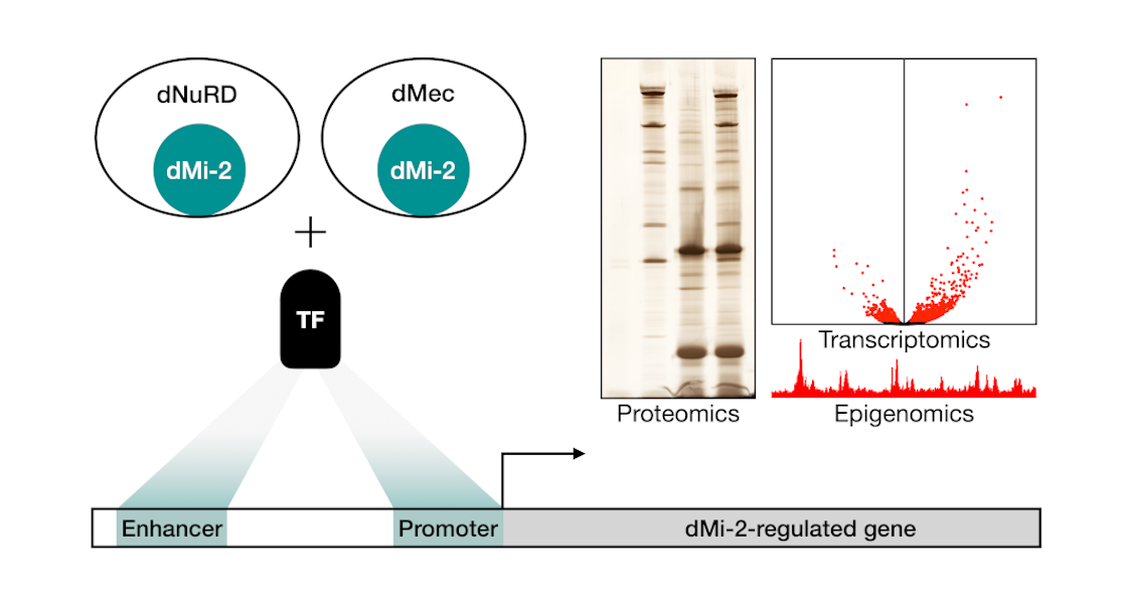
The Drosophila homolog of CHD4, dMi-2, is part of at least two distinct protein complexes: dNuRD and dMec. Both of these assemblies regulate gene expression and bind to chromatin. They are crucial for Drosophila development and differentiation of many specialised cell-types. Although we find that dMi-2 occupies distinct loci in the genome, none of the complex components have a known sequence preference in their DNA-binding.
This evokes the long standing question: How are chromatin regulators without sequence-specificity, like dMi-2, guided to their chromatin targets? We hypothesise that other DNA-binding proteins, that possess a sequence preference, tether dMi-2 to its target loci. To address this, we employ proteomic approaches (endogenous tagging using CRISPR/Cas9, immunoprecipitation, mass spectrometry, Western blot) to characterise novel dMi-2 interactors. Using next-generation sequencing (RNA-seq, ChIP-seq) we uncover genes and pathways that are regulated by dMi-2 and its associated factors.
In doing so we have found so far unknown cooperations of dMi-2 with transcription factors that function for example in steroid hormone response (Kreher et al., 2017), spermatogenesis (Kim et al., 2017) and hematopoiesis (Lenz et al., 2021).
This project was funded by the Deutsche Forschungsgemeinschaft (DFG, grants TRR81 A01 & BR2102/8).