Main Content
Research
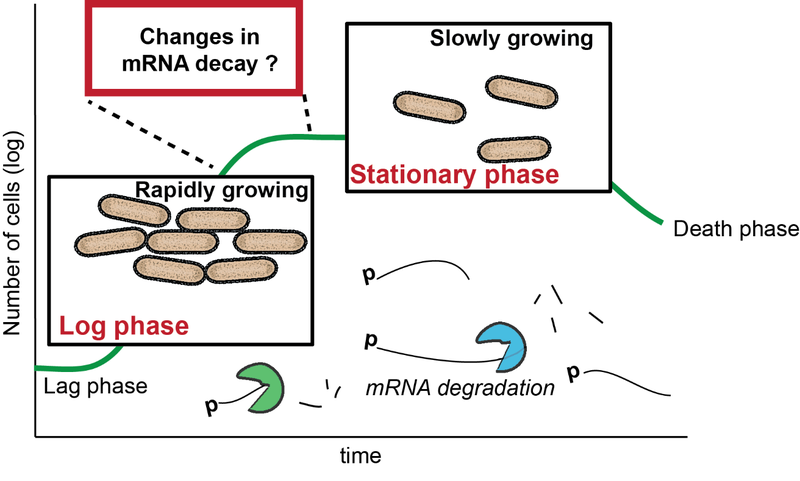
Working on the issue of mRNA processing has been done in exponentially growing cells, while only little is known about this essential process in transition and stationary growing cells. Thus, this research in the stationary phase is especially important due to the fact that under natural conditions bacteria are predominantly found in stationary growth. The gram-positive organism Bacillus subtilis serves as a model organism in many aspects and is already well-studied in many topics including competence, sporulation, and motility. The focus especially on B. subtilis is based on its ability to decide between different cell fates at the transition from exponential to stationary growth. Adjustment of protein levels and therefore availability and stability of mRNA is needed to ensure survival during adverse conditions. So far, a number of specific sigma factors like Sig H, Sig C, Sig D, and Sig B (σ38, Escherichia coli) as well as RNA-binding proteins, are known for the modification and thus regulation of mRNA levels in stationary growing B. subtilis cells. However, the involvement and regulation of ribonucleases (RNases) are largely unknown in this context, which definitively have a crucial role in regulating the protein levels and therefore, the energy of the cell. In B. subtilis 20 RNases are known of which some may be involved in this process.
Understanding these regulatory processes in stationary growing gram-positive and gram-negative bacteria will help to improve a better understanding of regulatory processes at the mRNA level in the bacterial cell.
Sources used:
Bechhofer, D. H. and Deutscher, M. P. “Bacterial ribonucleases and their roles in RNA metabolism.” Critical reviews in biochemistry and molecular biology vol. 54,3 (2019): 242-300. doi:10.1080/10409238.2019.1651816
Vargas-Blanco, D. A. and Shell, S. “Regulation of mRNA Stability During Bacterial Stress Responses.” Frontiers in Microbiology vol. 11 (2020): 2111. 10.3389/fmicb.2020.02111
Britton, R. A. et al. “Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis.” Journal of bacteriology vol. 184,17 (2002): 4881-90. doi:10.1128/JB.184.17.4881-4890.2002
Boylan, S. A. et al. “Transcription factor sigma B of Bacillus subtilis controls a large stationary-phase regulon.” Journal of bacteriology vol. 175,13 (1993): 3957-63. doi:10.1128/jb.175.13.3957-3963.1993
Jishage, M. and Ishihama, A. “Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of sigma 70 and sigma 38.” Journal of bacteriology vol. 177,23 (1995): 6832-5. doi:10.1128/jb.177.23.6832-6835.1995
Christopoulou, N. and Granneman, S. “The role of RNA-binding proteins in mediating adaptive responses in Gram-positive bacteria.” The FEBS journal vol. 289,7 (2022): 1746-1764. doi:10.1111/febs.15810
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. “Highly accurate protein structure prediction with AlphaFold.” Nature, 596(7873), (2021): 583-589.


