Main Content
Plant cryptochromes and photolyases
Cryptochromes are UV-A / blue light photoreceptors. They were first molecularly characterized in plants (1, 2) and subsequently found in many species from all kingdoms of life, including mammals. Cryptochromes are closely related to DNA photolyases and form with these enzymes the so-called cryptochrome / photolyase family (CPF, Figure 1).
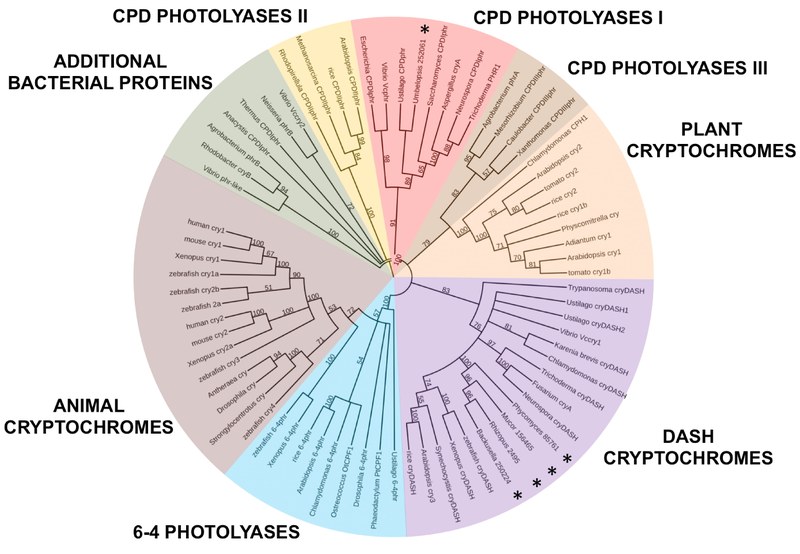
Cladogram representing the distribution and evolutionary relationships of members of the photolyase/cryptochrome family in a broad range of organisms. The eight subfamilies are indicated with different colors, following the classification of Chaves et al. (2011). Asterisks mark the position of members of the photolyase/cryptochrome family in mucoromycotina fungi. The phylogenetic tree is midpoint rooted and has been obtained using the NJ method with nodal support referred to 1000 pseudoreplicates. Only the topology is presented, and branches are not proportional to the amount of evolutionary change that has taken place along them. Bootstrap values above 50% are shown at the respective nodes. Bootstrap nodes under 50% have been collapsed. Taken from Tagua et al., (2015).
DNA photolyases use light energy to repair the two major lesions in DNA caused by UV-B. These damages are cyclobutane pyrimidine dimers (CPD) and (6-4) photoproducts (Figure 2; ref. 3).
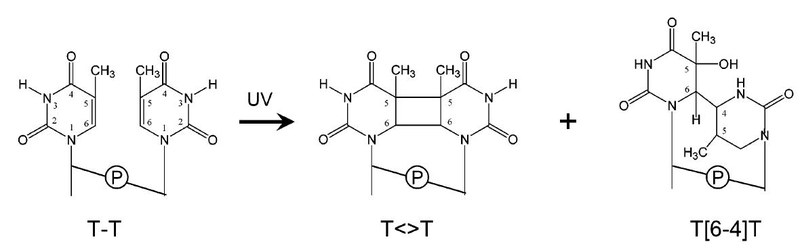
Shown are the two major UV-B photoproducts in DNA, cyclobutane pyrimidine dimers (CPD, T<>T) and (6-4)-photoproducts (T[6-4]T) between neighboring pyrimidines (here thymines). T<>T are the dominant lesions. Both lesions can be repaired by DNA-photolyases, enzymes each specific for T<>T or T[6-4]. Taken from Sancar, (2003).
Cryptochromes have no DNA repair activity, but high structural similarity to DNA photolyases in their N-terminal photolyase homologous region (PHR). In contrast to photolyases, cryptochromes usually have a C-terminal extension (CCE, Figure 3).
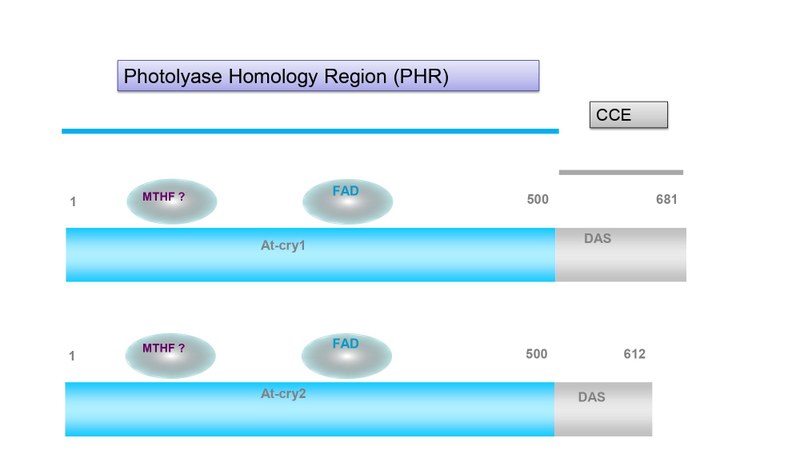
Plant cryptochromes contain an N-terminal photolyase homology region (PHR) of about 500 amino acid residues and the Cryptochrome C-terminal extensions (CCE). The PHR domain binds the FAD cofactor. Binding of methenyltetrahydrofolate (MTHF) as antenna chromophore is not confirmed by all studies. The CCEs in cryptochromes have variable lengths. Conserved is the so called DAS motif in plant cryptochromes consisting of aspartate (D), acidic (A) and serine (S) residues.
All members of CPF carry flavin adenine dinucleotide (FAD) as a cofactor. FAD must be completely reduced in photolyases for their catalytic activity. In cryptochromes, FAD is in the light-excited state in the semi-reduced form (ref. 4, 5). The main function of cryptochromes in plants is regulation of photoperiod flowering, deetiolation, and entrainment of the circadian clock.
Our current focus is on the activation and deactivation mechanism of cryptochromes. Thus, we were able to show that plant cryptochromes bind nucleotides such as ATP and thereby the photocycle is influenced with more active cryptochrome formed. Cryptochrome mutants that can no longer bind ATP are less biologically active than the wild type (6-8). Further work deals with the oligomerization of cryptochromes and its influence on the activity of these photoreceptors. Part of our work is dedicated to structure solution of photoreceptors as exemplified by Arabidopsis cry3 (Figure 4).
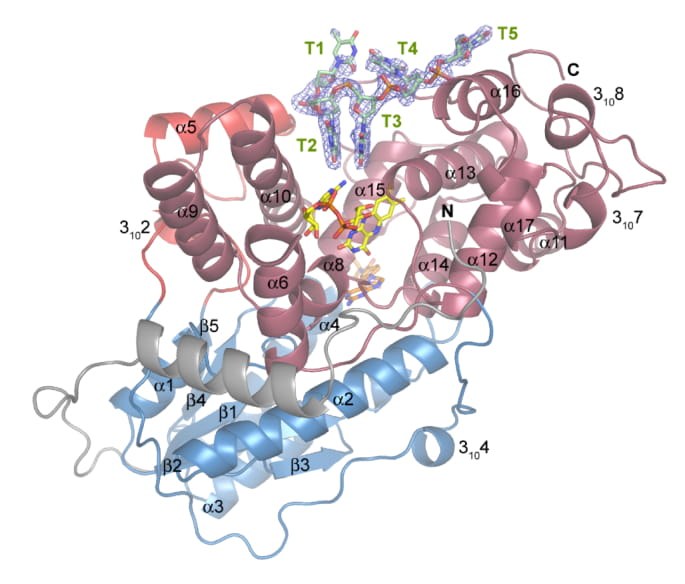
Ribbon model of complex A from A. thaliana cry3 and the repaired CPD damage in sticks representation with SIGMAA-weighted Fobs-Fcalc omit electron density (light blue, contoured at 2 σ) defining the oligonucleotide. The catalytic domain (dark red) of A. t. cry3 contains the catalytic FAD cofactor (yellow) and the antenna chromophore MTHF (orange) in the contact region to the antenna domain (blue). Nomenclature and definition of secondary structure elements are given in (Brudler R. et al. (2003) Identification of a new cryptochrome class: Structure, function, and evolution. Mol Cell 11:59-67.12). Taken from Pokorny et al. (2008).
Methodologically, this work is addressed by in vitro analyzes to structure elucidation of heterologously expressed proteins, as well as by genetic and photobiological analysis of Arabidopsis thaliana mutants and transgenic lines.
Team: Nils Niemann, Dennis Kock, Stephan Kiontke
Collaboration: Lars-Oliver Essen, Uwe Linne (University Marburg)
Funding: DFG Project BA 985/15-1
References:
- Batschauer A. (1993). A plant gene for photolyase: An enzyme catalyzing the repair of UV-light-induced DNA damage. Plant J. 4, 705-709.
- Ahmad M, Cashmore A (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166.
- Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue light photoreceptors. Chem. Rev. 103, 2203-2237.
- Banerjee R., Schleicher E., Meier S., Muñoz Viana R., Pokorny, R., Ahmad M., Bittl R., Batschauer A. (2007) The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem. 282, 14916-14922.
- Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Galland P, Bittl R, Ahmad M (2007) Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282, 9383-9391.
- Engelhard C., Wang X., Robles D., Moldt J., Essen L.-O., Batschauer A., Bittl R., Ahmad M. (2014). Cellular metabolites enhance light sensitivity of Arabidopsis cryptochrome through alternate electron transfer pathways. Plant Cell 26, 4519-4531.
- Orth C., Niemann N., Hennig L., Essen L.-O., Batschauer A. (2017) Hyperactivity of the Arabidopsis cryptochrome 1 (cry1) L407F mutant is caused by a structural alteration close to the cry1 ATP-binding site. J. Biol. Chem. 292, 12906-12920.
- Eckel M., Steinchen W.A., Batschauer A. (2018) ATP boosts lit state formation and activity of Arabidopsis cryptochrome 2. Plant J., 96, 389-403.
- Pokorny R., Klar T., Hennecke U., Carell T., Batschauer A., Essen L.-O. (2008) Recognition and repair of UV-lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc. Natl. Acad. Sci. USA 105, 21023-21027.