Main Content
Fungal Photoreceptors
Although fungi are not photosynthetically active, they nevertheless control many developmental processes and biochemical reactions by light. Light is used as a signal to inform the organism indirectly about changing environmental conditions, such as dryness, availability of nutrients, etc. Fungi have a considerable number of photoreceptors for this purpose. The by us intensively investigated biotrophic fungus Ustilago maydis, which induces the disease corn smut on corn plants (Figure 1), has 10 predicted light-sensitive proteins (Figure 2).

Figure 1. Symptoms on corn plants infected by Ustilago maydis.
U. maydis belongs to the class of basidiomycetes and is a facultative biotrophic fungus which infects Zea mays and its ancestor Teosinte.
The picture shows the formation of black teliospores on corn ears which is the final step in the biotrophic phase in the diphasic life cycle of U. maydis.
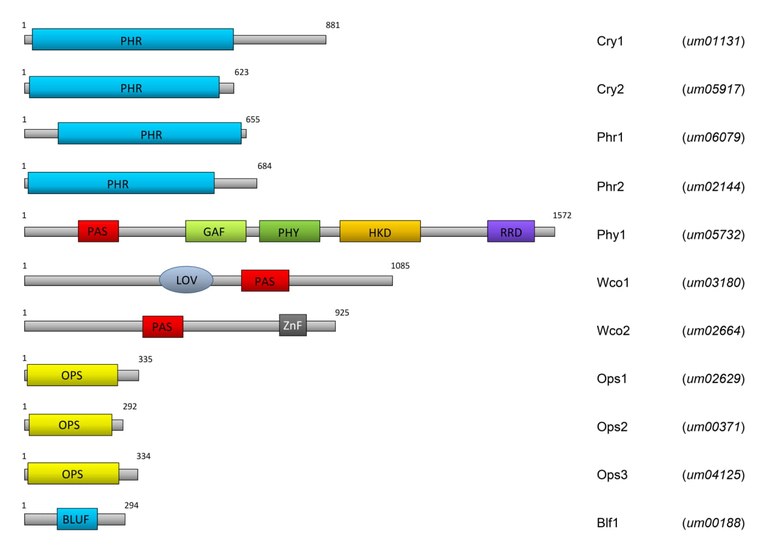
Abbreviations: PHR, Photolyase homology region; PAS, Per/Arndt/SIM domain; GAF, cGMP-specific phosphodiesterase/Anabaena adenylate cyclase/E. coli FhlA domain; PHY, phytochrome-specific domain; HKD, histidine kinase domain; RRD, response receiver domain; LOV, Light/Oxygen/Voltage domain; ZnF, zinc-finger domain; OPS, opsin domain; BLUF, blue light sensing using FAD domain. The numbers indicate the length of the proteins in amino acids. Taken from Brych et al. (2015).
Of these, some have been shown to act as photoreceptors (ref. 1). In Ustilago maydis there are four genes for members of the cryptochrome / photolyase family, one phytochrome (Phy1), one white collar 1 (Wco1) and one white collar 2 (Wco2) protein, three opsins and one so-called BLUF (Blue Light Sensing using Flavin) domain protein whose occurrence in fungi is unusual. As in other fungi, Wco1 and Wco2 dimerize, forming a so-called white collar complex (WCC), which controls the expression of numerous genes in U. maydis as an important blue light receptor (ref. 1). Two members of the CPF are canonical photolyases, two members are DASH cryptochromes. DASH (Drosophila, Arabidopsis, Synechocystis, Human) cryptochromes (cry-DASH) were first described for Synechocystis (ref. 2) and Arabidopsis (ref. 3), but later also detected in fungi and animals. Cry-DASH, in contrast to canonical photolyases, can only repair cyclobutane pyrimidine dimers in single-stranded DNA (refs. 4, 5) and are thus considered to be transitional forms between photolyases and cryptochromes. Like other mucorales, Phycomyces blakesleeanus has no canonical photolyases but only one cry-DASH (CryA). In collaboration with Luis Corrochano (University of Seville), we showed that CryA can also repair CPDs in double-stranded DNA (ref. 6) which is highly interesting also from an evolutionary point of view.
In collaboration with Ulrich Terpitz (University of Würzburg), we found that of the three opsins coded in U. maydis, two (Ops1, Ops2) bind retinal and act as green-light driven proton pumps (ref. 7). Ongoing work is devoted to the biological function of the photoreceptors of U. maydis and other fungi, as well as their molecular mechanism of action. Methodologically, these questions are addressed with recombinant proteins, with protein-protein interaction studies, spectroscopic analyzes, genetics and structural biology approaches as well as pathogenicity tests.
Team: Stephan Kiontke, Annika Brych, Jan Bräuer
Collaboration: Luis Corrochano, Javier Avalos, Carmen Limón (University of Seville), Ulrich Terpitz (University Würzburg), Stefan Rensing (University Marburg)
Funding: Collaborative Research Center (SFB) 987, DFG
References:
- Brych A., Mascarenhas J., Jaeger E., Charkiewicz E., Pokorny R., Bölker M., Doehlemann G., Batschauer A. (2015) White collar-1-induced photolyase expression contributes to UV-tolerance of Ustilago maydis. MicrobiologyOpen, doi: 10.1002/mbo3.322.
- Brudler, R., Hitomi, K., Daiyasu, H., Toh, H., Kucho, K., Ishiura, M., Kanehisa, M., Roberts, V.A., Todo, T., Tainer, J.A., and Getzoff, E.D. (2003). Identification of a new cryptochrome class. Structure, function, and evolution. Mol. Cell 11: 59-67.
- Kleine T. Lockhart, P., Batschauer A. (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 35, 93-103.
- Selby, C.P., and Sancar, A. (2006). A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 103: 17696-17700.
- Pokorny R., Klar T., Hennecke U., Carell T., Batschauer A., Essen L.-O. (2008) Recognition and repair of UV-lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc. Natl. Acad. Sci. USA 105, 21023-21027.
- Tagua V.G., Pausch M., Eckel M., Gutiérrez G., Miralles-Durána, A., Sanz C., Eslava A.P., Pokorny R., Corrochano L.M., Batschauer A. (2015) Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc. Natl. Acad. Sci. USA 112, 15130-15135.
- Panzer S., Brych A., Batschauer A., Terpitz U. (2019) Opsin 1 and opsin 2 of the corn smut fungus Ustilago maydis are green light-driven proton pumps. Front. Microbiol., doi: 10.3389/fmicb.2019.00735.